Gene
KWMTBOMO07686
Pre Gene Modal
BGIBMGA001278
Annotation
PREDICTED:_adenosine_deaminase-like_protein_[Papilio_xuthus]
Full name
Adenosine deaminase-like protein
Location in the cell
Cytoplasmic Reliability : 2.107
Sequence
CDS
ATGCTTCAGCTTCAAAGGTATTATGTCGACGCTGGCATATCTGATAAAACAAACACATTTCTCGACGAATTTCAAATAGGCGCTGGAGATACAAGAAATTTATCAGAATGTTTCCAAGTATTCAGTATTGCACATTCATTAACAAGCACGTCTGAAGCATTAGTAATGGCTACAGAACTGACACTCCAAGAATTTCAAGAAGATGGGTGTTGTTATATTGAGCTGAGAAGTACTCCACGTGACACACAGTACATAACAAAGAAGCAATATATTGATAGTATTATAAGGGCTATGGAAAAACCATCAATAAACCTCTCTATCATATCTAAATTGATTATATCAATAAATAGGGCATCCCAGCTACAAGAAGTGGAGGAAATAGCTGATATTGCAATAGAAAGGCATAAAATACACCCTGACACAGTTGTGGGAATAGAATTAAGTGGTAACCCAGCAGTGGGAAATTTTGGAGATTTTATACCAGCTTTGAATAGAGCAAGACAATCTGGACTCAAAGTAACACTGCATTGTGGAGAAGTATGCAACCCTGAGGAAGTACTGGAAATGTTAAATTTTAAACCTGAAAGAATTGGACACGGAGTTTGCATTCACCCTAAATATGGGGGGACGGAAGAAACATGGATAGCACTTTGCCAATCAAAGATACCTGTGGAAGTATGTCTGACAAGCAACGTGAATACAAAAATAACGCCCGATTACGAGTCGCATCATTTCAAAGAACTTATTGAAGCTGGATTGTCTGTTGTACTTTGTACTGATGACAAAGGAGTATTTGCGACATCTCTGTCTCAAGAATACAGGCTATGTGCTGAATCATTTGCACTTACACGGATTCAAGTAGCAAAATTATCTTTAGACGCAGCACAATGCATATTTTCAGAAGCGAATATTAGAGAGTTAATCATTGGAAAGATTTTAAAATTTATGAACGATTATACCTCATAA
Protein
MLQLQRYYVDAGISDKTNTFLDEFQIGAGDTRNLSECFQVFSIAHSLTSTSEALVMATELTLQEFQEDGCCYIELRSTPRDTQYITKKQYIDSIIRAMEKPSINLSIISKLIISINRASQLQEVEEIADIAIERHKIHPDTVVGIELSGNPAVGNFGDFIPALNRARQSGLKVTLHCGEVCNPEEVLEMLNFKPERIGHGVCIHPKYGGTEETWIALCQSKIPVEVCLTSNVNTKITPDYESHHFKELIEAGLSVVLCTDDKGVFATSLSQEYRLCAESFALTRIQVAKLSLDAAQCIFSEANIRELIIGKILKFMNDYTS
Summary
Description
Putative nucleoside deaminase. May catalyze the hydrolytic deamination of adenosine or some similar substrate and play a role in purine metabolism (By similarity).
Cofactor
Zn(2+)
Similarity
Belongs to the metallo-dependent hydrolases superfamily. Adenosine and AMP deaminases family.
Keywords
Complete proteome
Hydrolase
Metal-binding
Nucleotide metabolism
Reference proteome
Zinc
Feature
chain Adenosine deaminase-like protein
Uniprot
H9IVJ8
A0A2W1BVF5
A0A3S2NJ68
A0A2A4JSQ8
A0A2H1WS74
S4PSE9
+ More
A0A0L7KUS2 A0A2A3ESK7 A0A067QNU1 A0A2P8ZHE8 A0A0L7R7S9 A0A088AUM3 A0A154NXL3 E0VQH7 A0A0N0U494 A0A3B3DIV7 A0A026VYN0 A0A310SIB3 A0A3Q1IT58 H2LJB8 A0A3Q0SRV5 A0A3B5KW73 A0A1A8GST2 A0A1A8C5B7 A0A3P9HAX1 A0A0L0CPJ4 A0A3P9KSK6 A0A3Q3KU84 A0A2I4CXC3 A0A3Q3JER3 A0A2U9BVD3 E2A524 A0A3B4VIB8 A0A3B3C522 B4N997 A0A3B4H6G3 A0A3Q2WDK0 A0A3P8P100 A0A3P9B976 C3KH84 A0A3Q3WFS8 B4QXA0 A0A3B4CSY4 I3JP16 A0A3P8W1Y5 A0A084W2E7 I3JP17 A0A3B3RLS6 A0A1W4VQC0 W5K1V4 A0A151JYJ6 A0A3Q1EWM9 A0A1S3EQF0 B4JFM5 A0A3Q2ZGP1 B4LX54 Q9VHH7 A0A3Q3L7A8 K7J8B6 B3P1T6 B4KBY3 E2B4Y9 A0A3B5AAP2 B3M1M6 A0A1A8IX15 A0A3Q2QHS6 B4HKH9 A0A2D0RE82 G3P5W7 B4PU03 W5UEB0 Q4RS97 H3DF40 G3P5U9 H3C284 H3C6U2 G3P5U2 F4WE52 A0A182V966 Q7Q9M2 A0A087U549 A0A182LBG6 A0A2U9BX23 A0A2U9BVF8 A0A195CNQ0 G3H0V6 W5JPS1 G3WHK4 A0A2R9BXW2 Q4V9P6 A0A0K8UEA1 M3WIN5 A0A0A1X993 A0A3P4NI06 C1BKY0 A0A3Q0CHA7 A0A2U3WQR9 A0A3B0KFB5 A0A2P6TI82
A0A0L7KUS2 A0A2A3ESK7 A0A067QNU1 A0A2P8ZHE8 A0A0L7R7S9 A0A088AUM3 A0A154NXL3 E0VQH7 A0A0N0U494 A0A3B3DIV7 A0A026VYN0 A0A310SIB3 A0A3Q1IT58 H2LJB8 A0A3Q0SRV5 A0A3B5KW73 A0A1A8GST2 A0A1A8C5B7 A0A3P9HAX1 A0A0L0CPJ4 A0A3P9KSK6 A0A3Q3KU84 A0A2I4CXC3 A0A3Q3JER3 A0A2U9BVD3 E2A524 A0A3B4VIB8 A0A3B3C522 B4N997 A0A3B4H6G3 A0A3Q2WDK0 A0A3P8P100 A0A3P9B976 C3KH84 A0A3Q3WFS8 B4QXA0 A0A3B4CSY4 I3JP16 A0A3P8W1Y5 A0A084W2E7 I3JP17 A0A3B3RLS6 A0A1W4VQC0 W5K1V4 A0A151JYJ6 A0A3Q1EWM9 A0A1S3EQF0 B4JFM5 A0A3Q2ZGP1 B4LX54 Q9VHH7 A0A3Q3L7A8 K7J8B6 B3P1T6 B4KBY3 E2B4Y9 A0A3B5AAP2 B3M1M6 A0A1A8IX15 A0A3Q2QHS6 B4HKH9 A0A2D0RE82 G3P5W7 B4PU03 W5UEB0 Q4RS97 H3DF40 G3P5U9 H3C284 H3C6U2 G3P5U2 F4WE52 A0A182V966 Q7Q9M2 A0A087U549 A0A182LBG6 A0A2U9BX23 A0A2U9BVF8 A0A195CNQ0 G3H0V6 W5JPS1 G3WHK4 A0A2R9BXW2 Q4V9P6 A0A0K8UEA1 M3WIN5 A0A0A1X993 A0A3P4NI06 C1BKY0 A0A3Q0CHA7 A0A2U3WQR9 A0A3B0KFB5 A0A2P6TI82
EC Number
3.5.4.-
Pubmed
19121390
28756777
23622113
26227816
24845553
29403074
+ More
20566863 29451363 24508170 30249741 17554307 21551351 26108605 20798317 17994087 25186727 24487278 24438588 29240929 25329095 10731132 12537572 20075255 17550304 23127152 15496914 21719571 12364791 14747013 17210077 20966253 21804562 20920257 23761445 21709235 22722832 17975172 25830018 29178410
20566863 29451363 24508170 30249741 17554307 21551351 26108605 20798317 17994087 25186727 24487278 24438588 29240929 25329095 10731132 12537572 20075255 17550304 23127152 15496914 21719571 12364791 14747013 17210077 20966253 21804562 20920257 23761445 21709235 22722832 17975172 25830018 29178410
EMBL
BABH01001241
KZ149946
PZC76770.1
RSAL01000599
RVE41329.1
NWSH01000677
+ More
PCG74859.1 ODYU01010667 SOQ55925.1 GAIX01013908 JAA78652.1 JTDY01005569 KOB66875.1 KZ288192 PBC34262.1 KK853111 KDR11213.1 PYGN01000057 PSN55923.1 KQ414638 KOC66883.1 KQ434778 KZC04415.1 DS235430 EEB15633.1 KQ435850 KOX70951.1 KK107730 QOIP01000012 EZA47964.1 RLU15670.1 KQ759862 OAD62518.1 HAEC01005971 SBQ74048.1 HADZ01010095 SBP74036.1 JRES01000091 KNC34181.1 CP026252 AWP08248.1 GL436794 EFN71455.1 CH964232 EDW80530.1 BT082299 ACQ58006.1 CM000364 EDX13671.1 AERX01010904 ATLV01019609 KE525275 KFB44391.1 KQ981463 KYN41513.1 CH916369 EDV93506.1 CH940650 EDW66706.1 AE014297 BT031009 AAZX01003472 CH954181 EDV49824.1 CH933806 EDW15835.1 GL445681 EFN89253.1 CH902617 EDV43317.1 HAED01015465 SBR01910.1 CH480815 EDW42929.1 CM000160 EDW96614.1 JT413493 AHH40268.1 CAAE01015000 CAG08735.1 GL888102 EGI67490.1 AAAB01008900 EAA09431.4 KK118223 KFM72488.1 AWP08250.1 AWP08245.1 AWP08247.1 AWP08252.1 KQ977481 KYN02368.1 JH000098 EGV97216.1 ADMH02000689 ETN65308.1 AEFK01010432 AEFK01010433 AJFE02014749 BC096787 BC152246 GDHF01027421 JAI24893.1 AANG04001761 GBXI01006468 JAD07824.1 CYRY02019073 VCW96709.1 BT075259 ACO09683.1 OUUW01000008 SPP84416.1 LHPG02000015 PRW34001.1
PCG74859.1 ODYU01010667 SOQ55925.1 GAIX01013908 JAA78652.1 JTDY01005569 KOB66875.1 KZ288192 PBC34262.1 KK853111 KDR11213.1 PYGN01000057 PSN55923.1 KQ414638 KOC66883.1 KQ434778 KZC04415.1 DS235430 EEB15633.1 KQ435850 KOX70951.1 KK107730 QOIP01000012 EZA47964.1 RLU15670.1 KQ759862 OAD62518.1 HAEC01005971 SBQ74048.1 HADZ01010095 SBP74036.1 JRES01000091 KNC34181.1 CP026252 AWP08248.1 GL436794 EFN71455.1 CH964232 EDW80530.1 BT082299 ACQ58006.1 CM000364 EDX13671.1 AERX01010904 ATLV01019609 KE525275 KFB44391.1 KQ981463 KYN41513.1 CH916369 EDV93506.1 CH940650 EDW66706.1 AE014297 BT031009 AAZX01003472 CH954181 EDV49824.1 CH933806 EDW15835.1 GL445681 EFN89253.1 CH902617 EDV43317.1 HAED01015465 SBR01910.1 CH480815 EDW42929.1 CM000160 EDW96614.1 JT413493 AHH40268.1 CAAE01015000 CAG08735.1 GL888102 EGI67490.1 AAAB01008900 EAA09431.4 KK118223 KFM72488.1 AWP08250.1 AWP08245.1 AWP08247.1 AWP08252.1 KQ977481 KYN02368.1 JH000098 EGV97216.1 ADMH02000689 ETN65308.1 AEFK01010432 AEFK01010433 AJFE02014749 BC096787 BC152246 GDHF01027421 JAI24893.1 AANG04001761 GBXI01006468 JAD07824.1 CYRY02019073 VCW96709.1 BT075259 ACO09683.1 OUUW01000008 SPP84416.1 LHPG02000015 PRW34001.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000037510
UP000242457
UP000027135
+ More
UP000245037 UP000053825 UP000005203 UP000076502 UP000009046 UP000053105 UP000261560 UP000053097 UP000279307 UP000265040 UP000001038 UP000261340 UP000005226 UP000265200 UP000037069 UP000265180 UP000261640 UP000192220 UP000261600 UP000246464 UP000000311 UP000261420 UP000007798 UP000261460 UP000264840 UP000265100 UP000265160 UP000261620 UP000000304 UP000261440 UP000005207 UP000265120 UP000030765 UP000261540 UP000192221 UP000018467 UP000078541 UP000257200 UP000081671 UP000001070 UP000264800 UP000008792 UP000000803 UP000261660 UP000002358 UP000008711 UP000009192 UP000008237 UP000261400 UP000007801 UP000265000 UP000001292 UP000221080 UP000007635 UP000002282 UP000007303 UP000007755 UP000075903 UP000007062 UP000054359 UP000075882 UP000078542 UP000001075 UP000000673 UP000007648 UP000240080 UP000000437 UP000011712 UP000189706 UP000245340 UP000268350 UP000239899
UP000245037 UP000053825 UP000005203 UP000076502 UP000009046 UP000053105 UP000261560 UP000053097 UP000279307 UP000265040 UP000001038 UP000261340 UP000005226 UP000265200 UP000037069 UP000265180 UP000261640 UP000192220 UP000261600 UP000246464 UP000000311 UP000261420 UP000007798 UP000261460 UP000264840 UP000265100 UP000265160 UP000261620 UP000000304 UP000261440 UP000005207 UP000265120 UP000030765 UP000261540 UP000192221 UP000018467 UP000078541 UP000257200 UP000081671 UP000001070 UP000264800 UP000008792 UP000000803 UP000261660 UP000002358 UP000008711 UP000009192 UP000008237 UP000261400 UP000007801 UP000265000 UP000001292 UP000221080 UP000007635 UP000002282 UP000007303 UP000007755 UP000075903 UP000007062 UP000054359 UP000075882 UP000078542 UP000001075 UP000000673 UP000007648 UP000240080 UP000000437 UP000011712 UP000189706 UP000245340 UP000268350 UP000239899
Pfam
PF00962 A_deaminase
SUPFAM
SSF51556
SSF51556
ProteinModelPortal
H9IVJ8
A0A2W1BVF5
A0A3S2NJ68
A0A2A4JSQ8
A0A2H1WS74
S4PSE9
+ More
A0A0L7KUS2 A0A2A3ESK7 A0A067QNU1 A0A2P8ZHE8 A0A0L7R7S9 A0A088AUM3 A0A154NXL3 E0VQH7 A0A0N0U494 A0A3B3DIV7 A0A026VYN0 A0A310SIB3 A0A3Q1IT58 H2LJB8 A0A3Q0SRV5 A0A3B5KW73 A0A1A8GST2 A0A1A8C5B7 A0A3P9HAX1 A0A0L0CPJ4 A0A3P9KSK6 A0A3Q3KU84 A0A2I4CXC3 A0A3Q3JER3 A0A2U9BVD3 E2A524 A0A3B4VIB8 A0A3B3C522 B4N997 A0A3B4H6G3 A0A3Q2WDK0 A0A3P8P100 A0A3P9B976 C3KH84 A0A3Q3WFS8 B4QXA0 A0A3B4CSY4 I3JP16 A0A3P8W1Y5 A0A084W2E7 I3JP17 A0A3B3RLS6 A0A1W4VQC0 W5K1V4 A0A151JYJ6 A0A3Q1EWM9 A0A1S3EQF0 B4JFM5 A0A3Q2ZGP1 B4LX54 Q9VHH7 A0A3Q3L7A8 K7J8B6 B3P1T6 B4KBY3 E2B4Y9 A0A3B5AAP2 B3M1M6 A0A1A8IX15 A0A3Q2QHS6 B4HKH9 A0A2D0RE82 G3P5W7 B4PU03 W5UEB0 Q4RS97 H3DF40 G3P5U9 H3C284 H3C6U2 G3P5U2 F4WE52 A0A182V966 Q7Q9M2 A0A087U549 A0A182LBG6 A0A2U9BX23 A0A2U9BVF8 A0A195CNQ0 G3H0V6 W5JPS1 G3WHK4 A0A2R9BXW2 Q4V9P6 A0A0K8UEA1 M3WIN5 A0A0A1X993 A0A3P4NI06 C1BKY0 A0A3Q0CHA7 A0A2U3WQR9 A0A3B0KFB5 A0A2P6TI82
A0A0L7KUS2 A0A2A3ESK7 A0A067QNU1 A0A2P8ZHE8 A0A0L7R7S9 A0A088AUM3 A0A154NXL3 E0VQH7 A0A0N0U494 A0A3B3DIV7 A0A026VYN0 A0A310SIB3 A0A3Q1IT58 H2LJB8 A0A3Q0SRV5 A0A3B5KW73 A0A1A8GST2 A0A1A8C5B7 A0A3P9HAX1 A0A0L0CPJ4 A0A3P9KSK6 A0A3Q3KU84 A0A2I4CXC3 A0A3Q3JER3 A0A2U9BVD3 E2A524 A0A3B4VIB8 A0A3B3C522 B4N997 A0A3B4H6G3 A0A3Q2WDK0 A0A3P8P100 A0A3P9B976 C3KH84 A0A3Q3WFS8 B4QXA0 A0A3B4CSY4 I3JP16 A0A3P8W1Y5 A0A084W2E7 I3JP17 A0A3B3RLS6 A0A1W4VQC0 W5K1V4 A0A151JYJ6 A0A3Q1EWM9 A0A1S3EQF0 B4JFM5 A0A3Q2ZGP1 B4LX54 Q9VHH7 A0A3Q3L7A8 K7J8B6 B3P1T6 B4KBY3 E2B4Y9 A0A3B5AAP2 B3M1M6 A0A1A8IX15 A0A3Q2QHS6 B4HKH9 A0A2D0RE82 G3P5W7 B4PU03 W5UEB0 Q4RS97 H3DF40 G3P5U9 H3C284 H3C6U2 G3P5U2 F4WE52 A0A182V966 Q7Q9M2 A0A087U549 A0A182LBG6 A0A2U9BX23 A0A2U9BVF8 A0A195CNQ0 G3H0V6 W5JPS1 G3WHK4 A0A2R9BXW2 Q4V9P6 A0A0K8UEA1 M3WIN5 A0A0A1X993 A0A3P4NI06 C1BKY0 A0A3Q0CHA7 A0A2U3WQR9 A0A3B0KFB5 A0A2P6TI82
PDB
6IJP
E-value=2.75448e-45,
Score=457
Ontologies
PATHWAY
GO
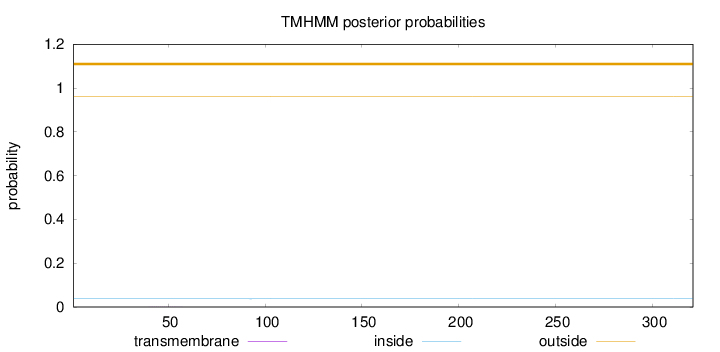
Topology
Length:
321
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00706999999999999
Exp number, first 60 AAs:
0.00437
Total prob of N-in:
0.03755
outside
1 - 321
Population Genetic Test Statistics
Pi
248.806451
Theta
201.93684
Tajima's D
0.899135
CLR
0.588687
CSRT
0.630468476576171
Interpretation
Uncertain
