Gene
KWMTBOMO07684 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA014605
Annotation
PREDICTED:_UPF0568_protein_C14orf166_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.147
Sequence
CDS
ATGACCAAAATTTTAATTTTGAAACAAAATGAAAAAATTGCTCTAGCAAAATATGCTAATATATATGATTTGCTAGTGGAGCCATGTGGCATTTTCATTAGACAAGAGCGCCCATATCTTGCTGCATCTCCTGATGGTCATCCAAATCCAGAGTTCTTTAACTGCGAAGATGAGAAGGAGTACAGAAGTGTGGTGCTGTGGTTAGAAGATCAAAAAATAAGGCATTACAAAATTGAGGACCGAGAAGGGTTGAGAAATATTGATTCCGATTTCTGGAAGGAGGCATACAATACGTATCAGAAAGATTTGGTGAGTCCAGCGGCAGAAGGGACGCTGAATGAACAACTCAATTGGCTATTGTCGTATGCGGTGCGGTTGGAATACGCGGATAATGTTGACAAATATAAAAATGCTAAAGTTGAAGAGAAAAAGAAAGCTCCCAATGTCGTGTCTTCTAATCCATTAGACAATCTGGACTTCTCAAGTCAAGCATTCAAAGCAGGTGTCAACAGAGTCTGTACAGTAGCGAGCATTGGTCCTCATCCAGATCCTAAAATAAGACTTGCTGCCTTAGCCAAAGTACTGAAAACTTCTCCGCATCCGGAACCCCTACAAACTGAAGTCAATTTAGTTAATCAACCCTCAGATGTGTTAAAGTTACTGTTTGTTCAGGATCTTAGAAATTTACAAACAAAAATTAATGAAGCTTTAGTTGCTGTCCAAACTGTTACTGCTGACCCTCGAACTGATACCAAACTCGGTCGAGTAGGAAGATAA
Protein
MTKILILKQNEKIALAKYANIYDLLVEPCGIFIRQERPYLAASPDGHPNPEFFNCEDEKEYRSVVLWLEDQKIRHYKIEDREGLRNIDSDFWKEAYNTYQKDLVSPAAEGTLNEQLNWLLSYAVRLEYADNVDKYKNAKVEEKKKAPNVVSSNPLDNLDFSSQAFKAGVNRVCTVASIGPHPDPKIRLAALAKVLKTSPHPEPLQTEVNLVNQPSDVLKLLFVQDLRNLQTKINEALVAVQTVTADPRTDTKLGRVGR
Summary
Uniprot
H9JYI5
A0A2H1WSE1
A0A2A4JS38
A0A0L7KUX5
A0A3S2LQ32
A0A212EKL5
+ More
A0A194PJ79 A0A194QZZ3 A0A2W1BNZ7 A0A1L8DAK3 A0A2J7R6Q2 A0A1B0CKB8 A0A1B6KTB4 A0A1B6FJ17 A0A067QWU6 A0A1B0D2S4 A0A1B6E935 A0A2R5LDU9 A0A0A9YSX6 A0A0K8SEH6 A0A161M9B6 E2J7G3 A0A0P4VSQ1 R4G5D5 A0A0V0G3U9 A0A069DR69 A0A023FT44 A0A023GC30 V5HG61 A0A1E1XS39 A0A1E1X4S7 A0A1S4ES89 A0A131Y3V1 R7VLV1 L7M662 A0A2R7W3N7 A0A224XVH6 A0A293N392 A0A1Z5L6Q8 A0A131XC10 A0A224YYB1 A0A3P8W4U5 A0A3Q2FMU4 A0A131YQS5 K1PKI9 A0A0P5SRW4 A0A1S3HNQ6 A0A1A8FEF1 A0A093PL73 A0A0P6B636 A0A3B3ZNR2 G3URA3 A0A1S3KHT1 A0A232ER27 V4B2H8 A0A1A8RVI9 A0A1A8DVX4 A0A1A8N1I8 Q90706 A0A1J1HRH1 A0A3B3BB21 A0A1A7YQ36 A0A2U9AVF3 A0A1A8IT62 A0A1V4KAB8 A0A1A8U4Q0 A0A226N989 A0A226P0B6 A0A1D5P7R1 G1NLZ3 A0A3B5K9T5 A0A3B5MUP3 A0A3P8W4S8 A0A3M0JIG2 U3JGV8 B5G0L3 A0A218UHB6 A0A3Q3LFX8 U5EZP8 A0A3B4WFU6 M4ALJ1 G3PLX0 A0A3S2Q0P0 A0A3Q3FK68 H2LFQ4 A0A0L8H738 A0A1U7S3G4 C1C3R2 A0A3B4ZJU4 A0A3Q1HUQ3 A0A3Q3QFS6 A0A3Q2W2D5 A0A3P8REC4 A0A3P9AWP3 A0A3Q1B0K3 A0A3P8TJG6 A0A3B4GM17 Q16JW3 A0A3P9IAJ0 A0A151PDZ6
A0A194PJ79 A0A194QZZ3 A0A2W1BNZ7 A0A1L8DAK3 A0A2J7R6Q2 A0A1B0CKB8 A0A1B6KTB4 A0A1B6FJ17 A0A067QWU6 A0A1B0D2S4 A0A1B6E935 A0A2R5LDU9 A0A0A9YSX6 A0A0K8SEH6 A0A161M9B6 E2J7G3 A0A0P4VSQ1 R4G5D5 A0A0V0G3U9 A0A069DR69 A0A023FT44 A0A023GC30 V5HG61 A0A1E1XS39 A0A1E1X4S7 A0A1S4ES89 A0A131Y3V1 R7VLV1 L7M662 A0A2R7W3N7 A0A224XVH6 A0A293N392 A0A1Z5L6Q8 A0A131XC10 A0A224YYB1 A0A3P8W4U5 A0A3Q2FMU4 A0A131YQS5 K1PKI9 A0A0P5SRW4 A0A1S3HNQ6 A0A1A8FEF1 A0A093PL73 A0A0P6B636 A0A3B3ZNR2 G3URA3 A0A1S3KHT1 A0A232ER27 V4B2H8 A0A1A8RVI9 A0A1A8DVX4 A0A1A8N1I8 Q90706 A0A1J1HRH1 A0A3B3BB21 A0A1A7YQ36 A0A2U9AVF3 A0A1A8IT62 A0A1V4KAB8 A0A1A8U4Q0 A0A226N989 A0A226P0B6 A0A1D5P7R1 G1NLZ3 A0A3B5K9T5 A0A3B5MUP3 A0A3P8W4S8 A0A3M0JIG2 U3JGV8 B5G0L3 A0A218UHB6 A0A3Q3LFX8 U5EZP8 A0A3B4WFU6 M4ALJ1 G3PLX0 A0A3S2Q0P0 A0A3Q3FK68 H2LFQ4 A0A0L8H738 A0A1U7S3G4 C1C3R2 A0A3B4ZJU4 A0A3Q1HUQ3 A0A3Q3QFS6 A0A3Q2W2D5 A0A3P8REC4 A0A3P9AWP3 A0A3Q1B0K3 A0A3P8TJG6 A0A3B4GM17 Q16JW3 A0A3P9IAJ0 A0A151PDZ6
Pubmed
19121390
26227816
22118469
26354079
28756777
24845553
+ More
25401762 26823975 27129103 26334808 25765539 29209593 28503490 23254933 25576852 28528879 28049606 28797301 24487278 26830274 22992520 25463417 20838655 28648823 29451363 24621616 15592404 21551351 17018643 23542700 17554307 25186727 17510324 22293439
25401762 26823975 27129103 26334808 25765539 29209593 28503490 23254933 25576852 28528879 28049606 28797301 24487278 26830274 22992520 25463417 20838655 28648823 29451363 24621616 15592404 21551351 17018643 23542700 17554307 25186727 17510324 22293439
EMBL
BABH01043858
BABH01043859
ODYU01010667
SOQ55927.1
NWSH01000677
PCG74857.1
+ More
JTDY01005569 KOB66876.1 RSAL01000599 RVE41327.1 AGBW02014249 OWR42020.1 KQ459606 KPI91140.1 KQ460947 KPJ10545.1 KZ149946 PZC76772.1 GFDF01010687 JAV03397.1 NEVH01006756 PNF36517.1 AJWK01015969 GEBQ01025319 JAT14658.1 GECZ01019635 JAS50134.1 KK853153 KDR10556.1 AJVK01023107 GEDC01023235 GEDC01002893 JAS14063.1 JAS34405.1 GGLE01003584 MBY07710.1 GBHO01016728 GBHO01009411 GBRD01014638 GBRD01014637 GDHC01021936 GDHC01021731 GDHC01015356 JAG26876.1 JAG34193.1 JAG51188.1 JAP96692.1 JAP96897.1 JAQ03273.1 GBRD01014636 JAG51190.1 GEMB01004112 JAR99150.1 HP429381 ADN29881.1 GDKW01001089 JAI55506.1 ACPB03011041 GAHY01000640 JAA76870.1 GECL01003370 JAP02754.1 GBGD01002683 JAC86206.1 GBBL01002766 JAC24554.1 GBBM01003999 JAC31419.1 GANP01007954 JAB76514.1 GFAA01001302 JAU02133.1 GFAC01004926 JAT94262.1 GEFM01003205 JAP72591.1 AMQN01003931 KB292234 ELU17935.1 GACK01006401 JAA58633.1 KK854240 PTY13600.1 GFTR01003986 JAW12440.1 GFWV01022857 MAA47584.1 GFJQ02004236 JAW02734.1 GEFH01003868 JAP64713.1 GFPF01011441 MAA22587.1 GEDV01006948 JAP81609.1 JH818712 EKC22168.1 GDIQ01177932 GDIQ01166791 GDIP01136209 GDIQ01005992 LRGB01000512 JAK73793.1 JAL67505.1 KZS18409.1 HAEB01011248 HAEC01005232 SBQ57775.1 KL670006 KFW77583.1 GDIP01019275 JAM84440.1 NNAY01002666 OXU20825.1 KB200701 ESP00602.1 HAEI01009982 HAEH01022347 SBS09314.1 HADZ01017791 HAEA01009597 SBQ38077.1 HAEF01021588 HAEG01007898 SBR62747.1 U46756 AAA87586.1 CVRI01000020 CRK90621.1 HADX01009773 SBP32005.1 CP026243 AWO95643.1 HAED01013243 HAEE01008316 SBQ99688.1 LSYS01003973 OPJ81301.1 HADY01018301 HAEJ01002613 SBS43070.1 MCFN01000133 OXB64068.1 AWGT02000251 OXB72819.1 AADN05000303 QRBI01000152 RMB98529.1 AGTO01021508 DQ215080 ACH44824.1 MUZQ01000302 OWK53177.1 GANO01000391 JAB59480.1 CM012438 RVE75301.1 KQ419106 KOF84575.1 BT081491 ACO51622.1 CH477989 EAT34579.1 AKHW03000474 KYO47228.1
JTDY01005569 KOB66876.1 RSAL01000599 RVE41327.1 AGBW02014249 OWR42020.1 KQ459606 KPI91140.1 KQ460947 KPJ10545.1 KZ149946 PZC76772.1 GFDF01010687 JAV03397.1 NEVH01006756 PNF36517.1 AJWK01015969 GEBQ01025319 JAT14658.1 GECZ01019635 JAS50134.1 KK853153 KDR10556.1 AJVK01023107 GEDC01023235 GEDC01002893 JAS14063.1 JAS34405.1 GGLE01003584 MBY07710.1 GBHO01016728 GBHO01009411 GBRD01014638 GBRD01014637 GDHC01021936 GDHC01021731 GDHC01015356 JAG26876.1 JAG34193.1 JAG51188.1 JAP96692.1 JAP96897.1 JAQ03273.1 GBRD01014636 JAG51190.1 GEMB01004112 JAR99150.1 HP429381 ADN29881.1 GDKW01001089 JAI55506.1 ACPB03011041 GAHY01000640 JAA76870.1 GECL01003370 JAP02754.1 GBGD01002683 JAC86206.1 GBBL01002766 JAC24554.1 GBBM01003999 JAC31419.1 GANP01007954 JAB76514.1 GFAA01001302 JAU02133.1 GFAC01004926 JAT94262.1 GEFM01003205 JAP72591.1 AMQN01003931 KB292234 ELU17935.1 GACK01006401 JAA58633.1 KK854240 PTY13600.1 GFTR01003986 JAW12440.1 GFWV01022857 MAA47584.1 GFJQ02004236 JAW02734.1 GEFH01003868 JAP64713.1 GFPF01011441 MAA22587.1 GEDV01006948 JAP81609.1 JH818712 EKC22168.1 GDIQ01177932 GDIQ01166791 GDIP01136209 GDIQ01005992 LRGB01000512 JAK73793.1 JAL67505.1 KZS18409.1 HAEB01011248 HAEC01005232 SBQ57775.1 KL670006 KFW77583.1 GDIP01019275 JAM84440.1 NNAY01002666 OXU20825.1 KB200701 ESP00602.1 HAEI01009982 HAEH01022347 SBS09314.1 HADZ01017791 HAEA01009597 SBQ38077.1 HAEF01021588 HAEG01007898 SBR62747.1 U46756 AAA87586.1 CVRI01000020 CRK90621.1 HADX01009773 SBP32005.1 CP026243 AWO95643.1 HAED01013243 HAEE01008316 SBQ99688.1 LSYS01003973 OPJ81301.1 HADY01018301 HAEJ01002613 SBS43070.1 MCFN01000133 OXB64068.1 AWGT02000251 OXB72819.1 AADN05000303 QRBI01000152 RMB98529.1 AGTO01021508 DQ215080 ACH44824.1 MUZQ01000302 OWK53177.1 GANO01000391 JAB59480.1 CM012438 RVE75301.1 KQ419106 KOF84575.1 BT081491 ACO51622.1 CH477989 EAT34579.1 AKHW03000474 KYO47228.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000283053
UP000007151
UP000053268
+ More
UP000053240 UP000235965 UP000092461 UP000027135 UP000092462 UP000015103 UP000079169 UP000014760 UP000265120 UP000265020 UP000005408 UP000076858 UP000085678 UP000053258 UP000261520 UP000001645 UP000215335 UP000030746 UP000183832 UP000261560 UP000246464 UP000190648 UP000198323 UP000198419 UP000000539 UP000005226 UP000261380 UP000269221 UP000016665 UP000197619 UP000261640 UP000261360 UP000002852 UP000007635 UP000261660 UP000001038 UP000053454 UP000189705 UP000261400 UP000265040 UP000261600 UP000264840 UP000265100 UP000265160 UP000257160 UP000265080 UP000261460 UP000008820 UP000265200 UP000050525
UP000053240 UP000235965 UP000092461 UP000027135 UP000092462 UP000015103 UP000079169 UP000014760 UP000265120 UP000265020 UP000005408 UP000076858 UP000085678 UP000053258 UP000261520 UP000001645 UP000215335 UP000030746 UP000183832 UP000261560 UP000246464 UP000190648 UP000198323 UP000198419 UP000000539 UP000005226 UP000261380 UP000269221 UP000016665 UP000197619 UP000261640 UP000261360 UP000002852 UP000007635 UP000261660 UP000001038 UP000053454 UP000189705 UP000261400 UP000265040 UP000261600 UP000264840 UP000265100 UP000265160 UP000257160 UP000265080 UP000261460 UP000008820 UP000265200 UP000050525
Pfam
PF10036 RLL
Interpro
IPR019265
RTRAF
ProteinModelPortal
H9JYI5
A0A2H1WSE1
A0A2A4JS38
A0A0L7KUX5
A0A3S2LQ32
A0A212EKL5
+ More
A0A194PJ79 A0A194QZZ3 A0A2W1BNZ7 A0A1L8DAK3 A0A2J7R6Q2 A0A1B0CKB8 A0A1B6KTB4 A0A1B6FJ17 A0A067QWU6 A0A1B0D2S4 A0A1B6E935 A0A2R5LDU9 A0A0A9YSX6 A0A0K8SEH6 A0A161M9B6 E2J7G3 A0A0P4VSQ1 R4G5D5 A0A0V0G3U9 A0A069DR69 A0A023FT44 A0A023GC30 V5HG61 A0A1E1XS39 A0A1E1X4S7 A0A1S4ES89 A0A131Y3V1 R7VLV1 L7M662 A0A2R7W3N7 A0A224XVH6 A0A293N392 A0A1Z5L6Q8 A0A131XC10 A0A224YYB1 A0A3P8W4U5 A0A3Q2FMU4 A0A131YQS5 K1PKI9 A0A0P5SRW4 A0A1S3HNQ6 A0A1A8FEF1 A0A093PL73 A0A0P6B636 A0A3B3ZNR2 G3URA3 A0A1S3KHT1 A0A232ER27 V4B2H8 A0A1A8RVI9 A0A1A8DVX4 A0A1A8N1I8 Q90706 A0A1J1HRH1 A0A3B3BB21 A0A1A7YQ36 A0A2U9AVF3 A0A1A8IT62 A0A1V4KAB8 A0A1A8U4Q0 A0A226N989 A0A226P0B6 A0A1D5P7R1 G1NLZ3 A0A3B5K9T5 A0A3B5MUP3 A0A3P8W4S8 A0A3M0JIG2 U3JGV8 B5G0L3 A0A218UHB6 A0A3Q3LFX8 U5EZP8 A0A3B4WFU6 M4ALJ1 G3PLX0 A0A3S2Q0P0 A0A3Q3FK68 H2LFQ4 A0A0L8H738 A0A1U7S3G4 C1C3R2 A0A3B4ZJU4 A0A3Q1HUQ3 A0A3Q3QFS6 A0A3Q2W2D5 A0A3P8REC4 A0A3P9AWP3 A0A3Q1B0K3 A0A3P8TJG6 A0A3B4GM17 Q16JW3 A0A3P9IAJ0 A0A151PDZ6
A0A194PJ79 A0A194QZZ3 A0A2W1BNZ7 A0A1L8DAK3 A0A2J7R6Q2 A0A1B0CKB8 A0A1B6KTB4 A0A1B6FJ17 A0A067QWU6 A0A1B0D2S4 A0A1B6E935 A0A2R5LDU9 A0A0A9YSX6 A0A0K8SEH6 A0A161M9B6 E2J7G3 A0A0P4VSQ1 R4G5D5 A0A0V0G3U9 A0A069DR69 A0A023FT44 A0A023GC30 V5HG61 A0A1E1XS39 A0A1E1X4S7 A0A1S4ES89 A0A131Y3V1 R7VLV1 L7M662 A0A2R7W3N7 A0A224XVH6 A0A293N392 A0A1Z5L6Q8 A0A131XC10 A0A224YYB1 A0A3P8W4U5 A0A3Q2FMU4 A0A131YQS5 K1PKI9 A0A0P5SRW4 A0A1S3HNQ6 A0A1A8FEF1 A0A093PL73 A0A0P6B636 A0A3B3ZNR2 G3URA3 A0A1S3KHT1 A0A232ER27 V4B2H8 A0A1A8RVI9 A0A1A8DVX4 A0A1A8N1I8 Q90706 A0A1J1HRH1 A0A3B3BB21 A0A1A7YQ36 A0A2U9AVF3 A0A1A8IT62 A0A1V4KAB8 A0A1A8U4Q0 A0A226N989 A0A226P0B6 A0A1D5P7R1 G1NLZ3 A0A3B5K9T5 A0A3B5MUP3 A0A3P8W4S8 A0A3M0JIG2 U3JGV8 B5G0L3 A0A218UHB6 A0A3Q3LFX8 U5EZP8 A0A3B4WFU6 M4ALJ1 G3PLX0 A0A3S2Q0P0 A0A3Q3FK68 H2LFQ4 A0A0L8H738 A0A1U7S3G4 C1C3R2 A0A3B4ZJU4 A0A3Q1HUQ3 A0A3Q3QFS6 A0A3Q2W2D5 A0A3P8REC4 A0A3P9AWP3 A0A3Q1B0K3 A0A3P8TJG6 A0A3B4GM17 Q16JW3 A0A3P9IAJ0 A0A151PDZ6
Ontologies
GO
PANTHER
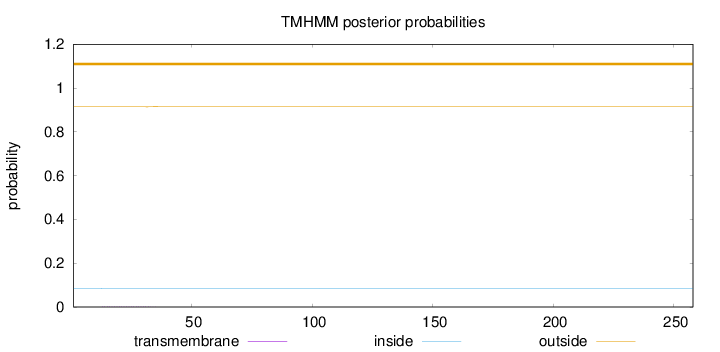
Topology
Length:
258
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0321
Exp number, first 60 AAs:
0.0321
Total prob of N-in:
0.08590
outside
1 - 258
Population Genetic Test Statistics
Pi
307.734152
Theta
192.83223
Tajima's D
2.022156
CLR
0.346646
CSRT
0.885855707214639
Interpretation
Uncertain
