Gene
KWMTBOMO07683
Pre Gene Modal
BGIBMGA001280
Annotation
PREDICTED:_dehydrodolichyl_diphosphate_synthase_isoform_X1_[Bombyx_mori]
Full name
Alkyl transferase
+ More
Dehydrodolichyl diphosphate synthase complex subunit DHDDS
Dehydrodolichyl diphosphate synthase complex subunit DHDDS
Alternative Name
Cis-isoprenyltransferase
Cis-prenyltransferase subunit hCIT
Epididymis tissue protein Li 189m
Cis-prenyltransferase subunit hCIT
Epididymis tissue protein Li 189m
Location in the cell
Cytoplasmic Reliability : 2.626
Sequence
CDS
ATGTCTTTGTGGATAAAAGAAAACTGTGTATCATTTTTCCAACTATTTTGTATCAAAGTAATAAAGACCGGCCGAGTACCCCAACACATAGCATTCATAATGGATGGAAACAGACGTTACGCGAAAAAAAATAGTGTCGACAAGAGTACTGGTCATCATAAAGGTTTTGACAAGCTTTCTGAAACCTTAAAGTGGTGCCTTGATTTAGGAATACCTGAGGTAACTGTTTATGCATTCAGCATAGAAAACTTTAAGAGAAGCAAAGAAGAAGTTGATGCACTTATGGAATTGGCTCGAGAAAAATTCCAAAATCTCTTGGATGAAATTGATCAAATTGATGAGTGGGGTGTACGAGTACATGTGGCTGGGCGACTCTCATTGTTACCGGAGCACTTGAGAAGTTTAGTTTCGAAGGTCATGCTCGCAACCAAAGATAACAACAGGCTTTGTTTGAACATTGCTTATGCCTACACAGGTCGTGATGAAATAAGCAGAGCAGCTTCACGTATAATTGACGGAGTCAAAGAAAATAAAATTTCTCCAGAAAATATAGATGAAGAACTGATTTCTCAATCACTGGATCTTGGAGAGCCTGAATTGCTTGTTAGAACATCAGGAGAAGTTAGGCTATCCGATTTTATGCTTTGGCAGATTTCGAATACCGTTCTCTATTTCACGGATGTATTGTGGCCAGAATTCACGATTTGGAATCTATTGGCGGCCATTATACATTTTCAGAGAAATGCACCTCCAATAAAAGCGGAAGACACCAATGAAACGGCGCACGAAAAGAAGGAATTTTTGAAATATTTAGAGCAAAGAAGAGCTCAAGAACTAACAAAATATATTAGTTGA
Protein
MSLWIKENCVSFFQLFCIKVIKTGRVPQHIAFIMDGNRRYAKKNSVDKSTGHHKGFDKLSETLKWCLDLGIPEVTVYAFSIENFKRSKEEVDALMELAREKFQNLLDEIDQIDEWGVRVHVAGRLSLLPEHLRSLVSKVMLATKDNNRLCLNIAYAYTGRDEISRAASRIIDGVKENKISPENIDEELISQSLDLGEPELLVRTSGEVRLSDFMLWQISNTVLYFTDVLWPEFTIWNLLAAIIHFQRNAPPIKAEDTNETAHEKKEFLKYLEQRRAQELTKYIS
Summary
Description
With NUS1, forms the dehydrodolichyl diphosphate synthase (DDS) complex, an essential component of the dolichol monophosphate (Dol-P) biosynthetic machinery. Both subunits contribute to enzymatic activity, i.e. condensation of multiple copies of isopentenyl pyrophosphate (IPP) to farnesyl pyrophosphate (FPP) to produce dehydrodolichyl diphosphate (Dedol-PP), a precursor of dolichol phosphate which is utilized as a sugar carrier in protein glycosylation in the endoplasmic reticulum (ER) (PubMed:25066056, PubMed:28842490). Regulates the glycosylation and stability of nascent NPC2, thereby promoting trafficking of LDL-derived cholesterol (PubMed:21572394).
Catalytic Activity
(2E,6E)-farnesyl diphosphate + n isopentenyl diphosphate = di-trans,poly-cis-polyprenyl diphosphate + n diphosphate
Cofactor
Mg(2+)
Biophysicochemical Properties
11.1 uM for isopentenyl diphosphate
0.68 uM for (2E,6E)-farnesyl diphosphate
0.68 uM for (2E,6E)-farnesyl diphosphate
Subunit
Forms an active dehydrodolichyl diphosphate synthase complex with NUS1 (PubMed:25066056, PubMed:28842490). Interacts with NPC2 (PubMed:21572394).
Similarity
Belongs to the UPP synthase family.
Keywords
Alternative splicing
Complete proteome
Disease mutation
Endoplasmic reticulum
Epilepsy
Lipid metabolism
Magnesium
Membrane
Mental retardation
Polymorphism
Reference proteome
Retinitis pigmentosa
Transferase
Feature
chain Alkyl transferase
splice variant In isoform 3.
sequence variant In DEDSM; unknown pathological significance.
splice variant In isoform 3.
sequence variant In DEDSM; unknown pathological significance.
Uniprot
A0A2A4JSU9
A0A2W1BYD1
A0A2H1WSA8
S4P8K6
A0A194PEL9
A0A212EKK9
+ More
E2B3T5 A0A0T6BAA2 A0A2J7QQ21 D6WVK3 A0A151WM42 A0A195CP43 E2BB83 A0A1W4WKN9 A0A195EU10 A0A067QWW1 A0A158NGB6 A0A195E5J8 F4WP20 A0A195B947 A0A1B6KAW1 A0A154PHN1 A0A1B6ELQ6 A0A1B6DEP7 A0A0L7R879 A0A1Y1N9T0 A0A232FDX9 A0A336MCK1 A0A336KQR1 A0A310SJE1 K7ISG5 A0A2A3E4X8 A0A088AJB7 A0A2M3ZCE7 A0A026WQ04 A0A2M3ZBI5 U5EUQ8 A0A084VTW0 A0A182J8A9 A0A182FQR3 S4R749 E9H940 A0A2M4BSY8 A0A2M4BSL9 A0A2M4BTU7 J3S8J7 A0A1D2N1S2 T1DN99 H9GFV7 U3JC76 A0A218UK33 A0A091EQK9 A0A1W7RI08 A0A182PRI8 A0A182K9Q2 A0A182QI47 A0A093PMZ7 A0A069DS28 R4FLJ6 A0A1U7UR60 A0A087VEC0 A0A2M4AKP0 F7I4W9 A0A3L8RX64 A0A2K6SQU5 I7GNQ9 Q177S1 L8IJJ2 A0A182N470 A0A182R6A9 A0A287AM35 A0A2I2YPT8 A0A2I3GV91 A0A2K5YZ47 A0A2R9AFF5 A0A2J8SIC4 G7NWT5 A0A1D5QZ83 A0A2H8TZA7 A0A1U7UG44 A0A151JMT0 W5JP25 Q86SQ9-2 B0W4D0 A0A182GER5 A0A341D6M2 A0A2U4BL69 A0A383ZUX4 A0A2Y9LKT1 U3CUJ0 A0A2I3RQZ6 C3YX74 A0A2Y9F084 A0A182YQZ3 B0W4C9 F6RWD2 M3WHN3 G1TCS2
E2B3T5 A0A0T6BAA2 A0A2J7QQ21 D6WVK3 A0A151WM42 A0A195CP43 E2BB83 A0A1W4WKN9 A0A195EU10 A0A067QWW1 A0A158NGB6 A0A195E5J8 F4WP20 A0A195B947 A0A1B6KAW1 A0A154PHN1 A0A1B6ELQ6 A0A1B6DEP7 A0A0L7R879 A0A1Y1N9T0 A0A232FDX9 A0A336MCK1 A0A336KQR1 A0A310SJE1 K7ISG5 A0A2A3E4X8 A0A088AJB7 A0A2M3ZCE7 A0A026WQ04 A0A2M3ZBI5 U5EUQ8 A0A084VTW0 A0A182J8A9 A0A182FQR3 S4R749 E9H940 A0A2M4BSY8 A0A2M4BSL9 A0A2M4BTU7 J3S8J7 A0A1D2N1S2 T1DN99 H9GFV7 U3JC76 A0A218UK33 A0A091EQK9 A0A1W7RI08 A0A182PRI8 A0A182K9Q2 A0A182QI47 A0A093PMZ7 A0A069DS28 R4FLJ6 A0A1U7UR60 A0A087VEC0 A0A2M4AKP0 F7I4W9 A0A3L8RX64 A0A2K6SQU5 I7GNQ9 Q177S1 L8IJJ2 A0A182N470 A0A182R6A9 A0A287AM35 A0A2I2YPT8 A0A2I3GV91 A0A2K5YZ47 A0A2R9AFF5 A0A2J8SIC4 G7NWT5 A0A1D5QZ83 A0A2H8TZA7 A0A1U7UG44 A0A151JMT0 W5JP25 Q86SQ9-2 B0W4D0 A0A182GER5 A0A341D6M2 A0A2U4BL69 A0A383ZUX4 A0A2Y9LKT1 U3CUJ0 A0A2I3RQZ6 C3YX74 A0A2Y9F084 A0A182YQZ3 B0W4C9 F6RWD2 M3WHN3 G1TCS2
EC Number
2.5.1.-
2.5.1.87
2.5.1.87
Pubmed
28756777
23622113
26354079
22118469
20798317
18362917
+ More
19820115 24845553 21347285 21719571 28004739 28648823 20075255 24508170 30249741 24438588 21292972 23025625 27289101 23758969 25727380 21881562 26358130 26334808 30282656 17194215 17510324 22751099 22398555 22722832 22002653 17431167 20920257 23761445 12591616 20736409 14702039 16710414 15489334 14652022 21572394 25066056 25944712 28842490 21295283 29100083 26483478 25243066 16136131 18563158 25244985 19393038 17975172 21993624
19820115 24845553 21347285 21719571 28004739 28648823 20075255 24508170 30249741 24438588 21292972 23025625 27289101 23758969 25727380 21881562 26358130 26334808 30282656 17194215 17510324 22751099 22398555 22722832 22002653 17431167 20920257 23761445 12591616 20736409 14702039 16710414 15489334 14652022 21572394 25066056 25944712 28842490 21295283 29100083 26483478 25243066 16136131 18563158 25244985 19393038 17975172 21993624
EMBL
NWSH01000677
PCG74856.1
KZ149946
PZC76773.1
ODYU01010667
SOQ55928.1
+ More
GAIX01004159 JAA88401.1 KQ459606 KPI91139.1 AGBW02014249 OWR42021.1 GL445407 EFN89604.1 LJIG01009070 KRT83815.1 NEVH01012087 PNF30688.1 KQ971357 EFA08584.1 KQ982944 KYQ48903.1 KQ977574 KYN01879.1 GL446946 EFN87056.1 KQ981965 KYN31733.1 KK853270 KDR09151.1 ADTU01015127 KQ979608 KYN20361.1 GL888243 EGI63951.1 KQ976542 KYM81063.1 GEBQ01031422 JAT08555.1 KQ434899 KZC10838.1 GECZ01030910 JAS38859.1 GEDC01029070 GEDC01013127 JAS08228.1 JAS24171.1 KQ414637 KOC66961.1 GEZM01011134 JAV93640.1 NNAY01000364 OXU28875.1 UFQT01000920 SSX28025.1 UFQS01000718 UFQT01000718 SSX06391.1 SSX26744.1 KQ760629 OAD59734.1 KZ288368 PBC26813.1 GGFM01005404 MBW26155.1 KK107139 QOIP01000003 EZA57766.1 RLU24743.1 GGFM01005079 MBW25830.1 GANO01003760 JAB56111.1 ATLV01016518 KE525092 KFB41404.1 GL732607 EFX71721.1 GGFJ01007038 MBW56179.1 GGFJ01006918 MBW56059.1 GGFJ01007037 MBW56178.1 JU174372 GBEX01000929 AFJ49898.1 JAI13631.1 LJIJ01000293 ODM99222.1 GAAZ01000677 GBKC01000961 GBKD01000609 JAA97266.1 JAG45109.1 JAG47009.1 AAWZ02028769 AGTO01010845 MUZQ01000250 OWK54139.1 KK718920 KFO60153.1 GDAY02000656 JAV50769.1 AXCN02000028 KL670084 KFW78158.1 GBGD01002363 JAC86526.1 ACPB03006512 GAHY01001358 JAA76152.1 KL490134 KFO10962.1 GGFK01008003 MBW41324.1 QUSF01000165 RLV88884.1 AB173304 BAE90366.1 CH477371 EAT42447.1 JH881135 ELR56278.1 AEMK02000045 CABD030001822 CABD030001823 CABD030001824 ADFV01067284 ADFV01067285 ADFV01067286 ADFV01067287 ADFV01067288 AJFE02029104 AJFE02029105 AJFE02029106 AJFE02029107 AJFE02029108 AJFE02029109 NDHI03003565 PNJ20539.1 AQIA01005243 AQIA01005244 CM001276 EHH49677.1 JSUE03000754 JSUE03000755 JSUE03000756 JSUE03000757 CM001253 EHH14488.1 GFXV01007799 MBW19604.1 KQ978889 KYN27644.1 ADMH02000517 ETN66137.1 AB090852 GU727641 AK023164 AK297134 AK316485 AL513365 CH471059 BC003643 BC004117 BC034152 DS231836 EDS33215.1 JXUM01058457 KQ562003 KXJ76936.1 GAMT01005223 GAMT01005222 GAMS01000019 GAMR01008903 GAMQ01004248 GAMP01007467 JAB06638.1 JAB23117.1 JAB25029.1 JAB37603.1 JAB45288.1 AACZ04072234 GG666562 EEN55092.1 EDS33214.1 AANG04003369 AAGW02062858
GAIX01004159 JAA88401.1 KQ459606 KPI91139.1 AGBW02014249 OWR42021.1 GL445407 EFN89604.1 LJIG01009070 KRT83815.1 NEVH01012087 PNF30688.1 KQ971357 EFA08584.1 KQ982944 KYQ48903.1 KQ977574 KYN01879.1 GL446946 EFN87056.1 KQ981965 KYN31733.1 KK853270 KDR09151.1 ADTU01015127 KQ979608 KYN20361.1 GL888243 EGI63951.1 KQ976542 KYM81063.1 GEBQ01031422 JAT08555.1 KQ434899 KZC10838.1 GECZ01030910 JAS38859.1 GEDC01029070 GEDC01013127 JAS08228.1 JAS24171.1 KQ414637 KOC66961.1 GEZM01011134 JAV93640.1 NNAY01000364 OXU28875.1 UFQT01000920 SSX28025.1 UFQS01000718 UFQT01000718 SSX06391.1 SSX26744.1 KQ760629 OAD59734.1 KZ288368 PBC26813.1 GGFM01005404 MBW26155.1 KK107139 QOIP01000003 EZA57766.1 RLU24743.1 GGFM01005079 MBW25830.1 GANO01003760 JAB56111.1 ATLV01016518 KE525092 KFB41404.1 GL732607 EFX71721.1 GGFJ01007038 MBW56179.1 GGFJ01006918 MBW56059.1 GGFJ01007037 MBW56178.1 JU174372 GBEX01000929 AFJ49898.1 JAI13631.1 LJIJ01000293 ODM99222.1 GAAZ01000677 GBKC01000961 GBKD01000609 JAA97266.1 JAG45109.1 JAG47009.1 AAWZ02028769 AGTO01010845 MUZQ01000250 OWK54139.1 KK718920 KFO60153.1 GDAY02000656 JAV50769.1 AXCN02000028 KL670084 KFW78158.1 GBGD01002363 JAC86526.1 ACPB03006512 GAHY01001358 JAA76152.1 KL490134 KFO10962.1 GGFK01008003 MBW41324.1 QUSF01000165 RLV88884.1 AB173304 BAE90366.1 CH477371 EAT42447.1 JH881135 ELR56278.1 AEMK02000045 CABD030001822 CABD030001823 CABD030001824 ADFV01067284 ADFV01067285 ADFV01067286 ADFV01067287 ADFV01067288 AJFE02029104 AJFE02029105 AJFE02029106 AJFE02029107 AJFE02029108 AJFE02029109 NDHI03003565 PNJ20539.1 AQIA01005243 AQIA01005244 CM001276 EHH49677.1 JSUE03000754 JSUE03000755 JSUE03000756 JSUE03000757 CM001253 EHH14488.1 GFXV01007799 MBW19604.1 KQ978889 KYN27644.1 ADMH02000517 ETN66137.1 AB090852 GU727641 AK023164 AK297134 AK316485 AL513365 CH471059 BC003643 BC004117 BC034152 DS231836 EDS33215.1 JXUM01058457 KQ562003 KXJ76936.1 GAMT01005223 GAMT01005222 GAMS01000019 GAMR01008903 GAMQ01004248 GAMP01007467 JAB06638.1 JAB23117.1 JAB25029.1 JAB37603.1 JAB45288.1 AACZ04072234 GG666562 EEN55092.1 EDS33214.1 AANG04003369 AAGW02062858
Proteomes
UP000218220
UP000053268
UP000007151
UP000008237
UP000235965
UP000007266
+ More
UP000075809 UP000078542 UP000192223 UP000078541 UP000027135 UP000005205 UP000078492 UP000007755 UP000078540 UP000076502 UP000053825 UP000215335 UP000002358 UP000242457 UP000005203 UP000053097 UP000279307 UP000030765 UP000075880 UP000069272 UP000245300 UP000000305 UP000094527 UP000001646 UP000016665 UP000197619 UP000052976 UP000075885 UP000075881 UP000075886 UP000053258 UP000015103 UP000189704 UP000008225 UP000276834 UP000233220 UP000008820 UP000075884 UP000075900 UP000008227 UP000001519 UP000001073 UP000233140 UP000240080 UP000009130 UP000233100 UP000006718 UP000000673 UP000005640 UP000002320 UP000069940 UP000249989 UP000252040 UP000245320 UP000261681 UP000248483 UP000002277 UP000001554 UP000248484 UP000076408 UP000009136 UP000011712 UP000001811
UP000075809 UP000078542 UP000192223 UP000078541 UP000027135 UP000005205 UP000078492 UP000007755 UP000078540 UP000076502 UP000053825 UP000215335 UP000002358 UP000242457 UP000005203 UP000053097 UP000279307 UP000030765 UP000075880 UP000069272 UP000245300 UP000000305 UP000094527 UP000001646 UP000016665 UP000197619 UP000052976 UP000075885 UP000075881 UP000075886 UP000053258 UP000015103 UP000189704 UP000008225 UP000276834 UP000233220 UP000008820 UP000075884 UP000075900 UP000008227 UP000001519 UP000001073 UP000233140 UP000240080 UP000009130 UP000233100 UP000006718 UP000000673 UP000005640 UP000002320 UP000069940 UP000249989 UP000252040 UP000245320 UP000261681 UP000248483 UP000002277 UP000001554 UP000248484 UP000076408 UP000009136 UP000011712 UP000001811
PRIDE
Pfam
PF01255 Prenyltransf
SUPFAM
SSF64005
SSF64005
Gene 3D
CDD
ProteinModelPortal
A0A2A4JSU9
A0A2W1BYD1
A0A2H1WSA8
S4P8K6
A0A194PEL9
A0A212EKK9
+ More
E2B3T5 A0A0T6BAA2 A0A2J7QQ21 D6WVK3 A0A151WM42 A0A195CP43 E2BB83 A0A1W4WKN9 A0A195EU10 A0A067QWW1 A0A158NGB6 A0A195E5J8 F4WP20 A0A195B947 A0A1B6KAW1 A0A154PHN1 A0A1B6ELQ6 A0A1B6DEP7 A0A0L7R879 A0A1Y1N9T0 A0A232FDX9 A0A336MCK1 A0A336KQR1 A0A310SJE1 K7ISG5 A0A2A3E4X8 A0A088AJB7 A0A2M3ZCE7 A0A026WQ04 A0A2M3ZBI5 U5EUQ8 A0A084VTW0 A0A182J8A9 A0A182FQR3 S4R749 E9H940 A0A2M4BSY8 A0A2M4BSL9 A0A2M4BTU7 J3S8J7 A0A1D2N1S2 T1DN99 H9GFV7 U3JC76 A0A218UK33 A0A091EQK9 A0A1W7RI08 A0A182PRI8 A0A182K9Q2 A0A182QI47 A0A093PMZ7 A0A069DS28 R4FLJ6 A0A1U7UR60 A0A087VEC0 A0A2M4AKP0 F7I4W9 A0A3L8RX64 A0A2K6SQU5 I7GNQ9 Q177S1 L8IJJ2 A0A182N470 A0A182R6A9 A0A287AM35 A0A2I2YPT8 A0A2I3GV91 A0A2K5YZ47 A0A2R9AFF5 A0A2J8SIC4 G7NWT5 A0A1D5QZ83 A0A2H8TZA7 A0A1U7UG44 A0A151JMT0 W5JP25 Q86SQ9-2 B0W4D0 A0A182GER5 A0A341D6M2 A0A2U4BL69 A0A383ZUX4 A0A2Y9LKT1 U3CUJ0 A0A2I3RQZ6 C3YX74 A0A2Y9F084 A0A182YQZ3 B0W4C9 F6RWD2 M3WHN3 G1TCS2
E2B3T5 A0A0T6BAA2 A0A2J7QQ21 D6WVK3 A0A151WM42 A0A195CP43 E2BB83 A0A1W4WKN9 A0A195EU10 A0A067QWW1 A0A158NGB6 A0A195E5J8 F4WP20 A0A195B947 A0A1B6KAW1 A0A154PHN1 A0A1B6ELQ6 A0A1B6DEP7 A0A0L7R879 A0A1Y1N9T0 A0A232FDX9 A0A336MCK1 A0A336KQR1 A0A310SJE1 K7ISG5 A0A2A3E4X8 A0A088AJB7 A0A2M3ZCE7 A0A026WQ04 A0A2M3ZBI5 U5EUQ8 A0A084VTW0 A0A182J8A9 A0A182FQR3 S4R749 E9H940 A0A2M4BSY8 A0A2M4BSL9 A0A2M4BTU7 J3S8J7 A0A1D2N1S2 T1DN99 H9GFV7 U3JC76 A0A218UK33 A0A091EQK9 A0A1W7RI08 A0A182PRI8 A0A182K9Q2 A0A182QI47 A0A093PMZ7 A0A069DS28 R4FLJ6 A0A1U7UR60 A0A087VEC0 A0A2M4AKP0 F7I4W9 A0A3L8RX64 A0A2K6SQU5 I7GNQ9 Q177S1 L8IJJ2 A0A182N470 A0A182R6A9 A0A287AM35 A0A2I2YPT8 A0A2I3GV91 A0A2K5YZ47 A0A2R9AFF5 A0A2J8SIC4 G7NWT5 A0A1D5QZ83 A0A2H8TZA7 A0A1U7UG44 A0A151JMT0 W5JP25 Q86SQ9-2 B0W4D0 A0A182GER5 A0A341D6M2 A0A2U4BL69 A0A383ZUX4 A0A2Y9LKT1 U3CUJ0 A0A2I3RQZ6 C3YX74 A0A2Y9F084 A0A182YQZ3 B0W4C9 F6RWD2 M3WHN3 G1TCS2
PDB
5KH5
E-value=9.85823e-40,
Score=409
Ontologies
GO
PANTHER
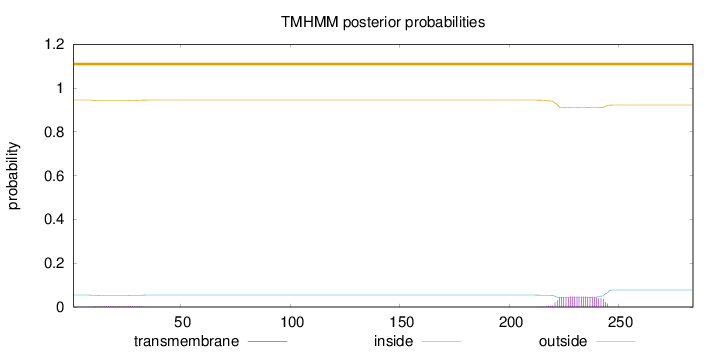
Topology
Subcellular location
Endoplasmic reticulum membrane
Length:
284
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.14557
Exp number, first 60 AAs:
0.07633
Total prob of N-in:
0.05492
outside
1 - 284
Population Genetic Test Statistics
Pi
179.810955
Theta
156.919098
Tajima's D
0.696144
CLR
0.0671
CSRT
0.56097195140243
Interpretation
Uncertain
