Gene
KWMTBOMO07679
Pre Gene Modal
BGIBMGA001281
Annotation
PREDICTED:_gem-associated_protein_5-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.731 Nuclear Reliability : 1.738
Sequence
CDS
ATGTCCTGTTCATCACATCTGCTGCTACATGTTGCTGTTGGTACTAAGCAAGGGGTTATATTTGTCTTGGATCTGAATGGTCAGGGAAAAACTATTTACAAAGTTCGAGGTCAAGATGATGAGATAATGACTGTAAGTTGGTGCCCACAGTATGAAGTTGTTATAAAAAAATTGTTGAAGGAATCGGATAAAAGACCCAGTGTATCTGAAAGACTACATAAGATAAGAAACGAACCAGAACCTGATGAAATTGAAGATTCAAATCCGAAGAGTAGTGAAAATTTAGAAGCGTCTGGCATGGTTAAGAGCCTTCCTGAAGATAGTTTTGATGAATCTGTCACTGTAGAAGATGATATGTTTGACATTTATAAAGATCACGAAGCTGATGAATTCGGACACAAAAAATATCAACCAGAAGAAATACGTGTAAAAATAAAACCAGATTTGAAACAAGAAGATTATTTAGCTGAATGTTTGAAATTGAAGGAACAAATTTTACGAAATAAAAATGAAACAGAAGAATCTATAGAGTCATTAGTTGAGGCCCTAGACAAAACTCACGTCAGTAATGATGAATCACATAATGATGTATTAGATGATAAAAACAAAGATGATCTAGTACAAGGCAGTGATCATGTGCATAAGCATCTTTTAGCTACAATTGGAAAATTGGGTGGAGTAAGAATATGGTCGAAAACCGGCAAGTTGATAGCAAGTTGTGCAGTGCCACAGAACAAGAACATGAAAATAAAATCACCCAGCTGGCCTTCATTATTATGGTATAAATCGAATATGCTACTCATCGGAGACAACAGAAGTCAGTTGCTGGAATGCAATCCATTGGTAATTGATTGCAAGAATAAATTAGACTGGAAAATAGTCCACAGTCTACATAAGCGCGGATTGTATTCCTTGACAACGAATGCTCCAAGAGTTCAGACAGAAGCTGACAAACACAAAATCTGGTCAGTGTGGTCTACGTCACAGGACAGAAATATCATAGAATATTGTGTCGAGAAACGGGAGAAAGTAGCCGTACATGCCACCTGTGGACGTTTTGTGTATTGCATACGGAGCTGCCCCTATGACGCGGGCAAAATAGCAATAAGCGTTGGAGATGGTACAGTTCGCATCTGGGAAACCAATACACTGGATCATGATGATACTAAACTTGCTGCTGGCACCATTACTGCCTACTGGCAGAATGTTCAAGGAAGAGTTTTGACATTAGCCTGGCATCCGACAAAAGAAAATTGGCTAGCCTTCAGCACTGCCGAGTCCAGAGTGGGGATCCTGGACACGAGCGGTCGCGGCGCCCGCGCGCTGGTGCCGGGGCTGGCGGGCGGCGTGTACTCGCTGTGCTGGGGGCTGCGCTCGCACCTGTACGCCTGCGCCTCCGGACAACTCGTCGTGTACGACGCCGCCAGACCGGACCAACCCCCAGTACCAGTAGGAGTGGAGTTCGAAGGGCAAAAGTGGGAAATTAGTTCTATTAGCTGGAGTAGTCGCGGCTTTCTGGTGGGCAGTAACTCTGGTGGTGTTGCAGTACTCACGCACGAATTACCTCATACAGTCACGGCTGCTGTGTTTGCATTCAGTAAAATGATTCATTCAATAGATTGGCACCCTCAACAAACTTCTAATTCAAATGAAGAATCTCCTTTAAAAGATATGATTGCGGTTTGTTCATTGGACAATCAAAACACAATTATAATACTCGAGTATAGTGATAAAGGCGATCACGGACAGCTGACAACATGGAAAACGTTACGAGGCCACAAGGGAACCGTGTTCGAAGCGTCTTGGAACCCGCATCATGACGATCTGCTGCTGTCCACGTCTTCCGACGCCACAGTGCGGGTGTGGGCGGCGTCTAGTGGCGCCTGCGTGTCTGTGTTCGACGGGCACATGGCGCAGAGCGCGCTGGGCGCGGCGTGGAGCGCCTACCCGCAGCTCGCCACCAAGGCGCTGTCCGGGGGAGGAGACCACACGCTGAGGCTGTGGGACATGAACGACTTCCCCGCCGAAGCATACGACGGTCAGTTCATCACGAATCAATATTAG
Protein
MSCSSHLLLHVAVGTKQGVIFVLDLNGQGKTIYKVRGQDDEIMTVSWCPQYEVVIKKLLKESDKRPSVSERLHKIRNEPEPDEIEDSNPKSSENLEASGMVKSLPEDSFDESVTVEDDMFDIYKDHEADEFGHKKYQPEEIRVKIKPDLKQEDYLAECLKLKEQILRNKNETEESIESLVEALDKTHVSNDESHNDVLDDKNKDDLVQGSDHVHKHLLATIGKLGGVRIWSKTGKLIASCAVPQNKNMKIKSPSWPSLLWYKSNMLLIGDNRSQLLECNPLVIDCKNKLDWKIVHSLHKRGLYSLTTNAPRVQTEADKHKIWSVWSTSQDRNIIEYCVEKREKVAVHATCGRFVYCIRSCPYDAGKIAISVGDGTVRIWETNTLDHDDTKLAAGTITAYWQNVQGRVLTLAWHPTKENWLAFSTAESRVGILDTSGRGARALVPGLAGGVYSLCWGLRSHLYACASGQLVVYDAARPDQPPVPVGVEFEGQKWEISSISWSSRGFLVGSNSGGVAVLTHELPHTVTAAVFAFSKMIHSIDWHPQQTSNSNEESPLKDMIAVCSLDNQNTIIILEYSDKGDHGQLTTWKTLRGHKGTVFEASWNPHHDDLLLSTSSDATVRVWAASSGACVSVFDGHMAQSALGAAWSAYPQLATKALSGGGDHTLRLWDMNDFPAEAYDGQFITNQY
Summary
Uniprot
A0A3S2LFK8
A0A212EKL3
A0A2W1BT15
A0A2A4JTS7
A0A2H1WS83
A0A194PD27
+ More
A0A194QYK5 A0A154PRA4 A0A0P6EJQ9 A0A0P5M057 A0A0N8E160 A0A0P6IQX4 A0A0P6AW20 A0A0P5T8N9 A0A0P5Y6Y2 A0A0P5YU19 K7J2X6 A0A232FEJ5 A0A2A3ECB7 E2B371 A0A088A7B4 A0A0C9R3K7 A0A310SU74 A0A0M8ZSJ0 A0A195E488 A0A026WZX8 A0A151X220 A0A195FIX4 F4W6G0 A0A158P272 A0A195AVJ2 K1QI76 T1JBR0 D1ZZV0 A0A2T7PMS1 A0A091HME1 J9LLJ4 A0A1W2WMP3 A0A091RW82 A0A1S3HRG7 V9K8J8 F6ZHD7 F6WUT8 A0A1S3K5E0 A0A091K569 A0A091QBN7 U3K106 A0A093GJL0 A0A099ZQ50 A0A091IV06 A0A0Q3U3C8 A0A1Y1ML03 W5PKH7 A0A3L8SK07 A0A091V691 A0A091LVX5 A0A094LA71
A0A194QYK5 A0A154PRA4 A0A0P6EJQ9 A0A0P5M057 A0A0N8E160 A0A0P6IQX4 A0A0P6AW20 A0A0P5T8N9 A0A0P5Y6Y2 A0A0P5YU19 K7J2X6 A0A232FEJ5 A0A2A3ECB7 E2B371 A0A088A7B4 A0A0C9R3K7 A0A310SU74 A0A0M8ZSJ0 A0A195E488 A0A026WZX8 A0A151X220 A0A195FIX4 F4W6G0 A0A158P272 A0A195AVJ2 K1QI76 T1JBR0 D1ZZV0 A0A2T7PMS1 A0A091HME1 J9LLJ4 A0A1W2WMP3 A0A091RW82 A0A1S3HRG7 V9K8J8 F6ZHD7 F6WUT8 A0A1S3K5E0 A0A091K569 A0A091QBN7 U3K106 A0A093GJL0 A0A099ZQ50 A0A091IV06 A0A0Q3U3C8 A0A1Y1ML03 W5PKH7 A0A3L8SK07 A0A091V691 A0A091LVX5 A0A094LA71
Pubmed
EMBL
RSAL01000155
RVE45644.1
AGBW02014249
OWR42023.1
KZ149946
PZC76775.1
+ More
NWSH01000677 PCG74873.1 ODYU01010667 SOQ55930.1 KQ459606 KPI91137.1 KQ460947 KPJ10542.1 KQ435012 KZC13808.1 GDIQ01068643 JAN26094.1 GDIQ01171650 JAK80075.1 GDIQ01071988 JAN22749.1 GDIQ01001527 JAN93210.1 GDIP01025750 JAM77965.1 GDIP01146322 GDIP01135873 GDIP01135872 GDIP01096839 GDIP01096838 GDIP01074038 LRGB01002993 JAL67841.1 KZS05197.1 GDIP01075328 JAM28387.1 GDIP01066733 JAM36982.1 NNAY01000356 OXU28940.1 KZ288296 PBC28932.1 GL445305 EFN89853.1 GBYB01002530 JAG72297.1 KQ759878 OAD62256.1 KQ435918 KOX68733.1 KQ979657 KYN19901.1 KK107054 EZA61582.1 KQ982580 KYQ54459.1 KQ981523 KYN40353.1 GL887707 EGI70295.1 ADTU01007002 ADTU01007003 KQ976731 KYM76201.1 JH816294 EKC33488.1 JH432018 KQ971338 EFA02425.1 PZQS01000003 PVD34720.1 KL217541 KFO96364.1 ABLF02040311 KK935207 KFQ46840.1 JW861896 AFO94413.1 EAAA01001438 KK546863 KFP32612.1 KK659118 KFQ06989.1 AGTO01010809 KL216598 KFV70450.1 KL895988 KGL83083.1 KK501093 KFP12534.1 LMAW01000148 KQL60424.1 GEZM01031052 JAV85085.1 AMGL01093654 QUSF01000017 RLW03061.1 KL410715 KFQ98588.1 KK509422 KFP62542.1 KL263879 KFZ60964.1
NWSH01000677 PCG74873.1 ODYU01010667 SOQ55930.1 KQ459606 KPI91137.1 KQ460947 KPJ10542.1 KQ435012 KZC13808.1 GDIQ01068643 JAN26094.1 GDIQ01171650 JAK80075.1 GDIQ01071988 JAN22749.1 GDIQ01001527 JAN93210.1 GDIP01025750 JAM77965.1 GDIP01146322 GDIP01135873 GDIP01135872 GDIP01096839 GDIP01096838 GDIP01074038 LRGB01002993 JAL67841.1 KZS05197.1 GDIP01075328 JAM28387.1 GDIP01066733 JAM36982.1 NNAY01000356 OXU28940.1 KZ288296 PBC28932.1 GL445305 EFN89853.1 GBYB01002530 JAG72297.1 KQ759878 OAD62256.1 KQ435918 KOX68733.1 KQ979657 KYN19901.1 KK107054 EZA61582.1 KQ982580 KYQ54459.1 KQ981523 KYN40353.1 GL887707 EGI70295.1 ADTU01007002 ADTU01007003 KQ976731 KYM76201.1 JH816294 EKC33488.1 JH432018 KQ971338 EFA02425.1 PZQS01000003 PVD34720.1 KL217541 KFO96364.1 ABLF02040311 KK935207 KFQ46840.1 JW861896 AFO94413.1 EAAA01001438 KK546863 KFP32612.1 KK659118 KFQ06989.1 AGTO01010809 KL216598 KFV70450.1 KL895988 KGL83083.1 KK501093 KFP12534.1 LMAW01000148 KQL60424.1 GEZM01031052 JAV85085.1 AMGL01093654 QUSF01000017 RLW03061.1 KL410715 KFQ98588.1 KK509422 KFP62542.1 KL263879 KFZ60964.1
Proteomes
UP000283053
UP000007151
UP000218220
UP000053268
UP000053240
UP000076502
+ More
UP000076858 UP000002358 UP000215335 UP000242457 UP000008237 UP000005203 UP000053105 UP000078492 UP000053097 UP000075809 UP000078541 UP000007755 UP000005205 UP000078540 UP000005408 UP000007266 UP000245119 UP000054308 UP000007819 UP000085678 UP000008144 UP000016665 UP000053875 UP000053641 UP000053119 UP000051836 UP000002356 UP000276834 UP000053283
UP000076858 UP000002358 UP000215335 UP000242457 UP000008237 UP000005203 UP000053105 UP000078492 UP000053097 UP000075809 UP000078541 UP000007755 UP000005205 UP000078540 UP000005408 UP000007266 UP000245119 UP000054308 UP000007819 UP000085678 UP000008144 UP000016665 UP000053875 UP000053641 UP000053119 UP000051836 UP000002356 UP000276834 UP000053283
Interpro
IPR017986
WD40_repeat_dom
+ More
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR020472 G-protein_beta_WD-40_rep
IPR013783 Ig-like_fold
IPR022113 TMEM131-like_dom
IPR003961 FN3_dom
IPR036116 FN3_sf
IPR039877 TMEM131-like
IPR011041 Quinoprot_gluc/sorb_DH
IPR024977 Apc4_WD40_dom
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR020472 G-protein_beta_WD-40_rep
IPR013783 Ig-like_fold
IPR022113 TMEM131-like_dom
IPR003961 FN3_dom
IPR036116 FN3_sf
IPR039877 TMEM131-like
IPR011041 Quinoprot_gluc/sorb_DH
IPR024977 Apc4_WD40_dom
Gene 3D
CDD
ProteinModelPortal
A0A3S2LFK8
A0A212EKL3
A0A2W1BT15
A0A2A4JTS7
A0A2H1WS83
A0A194PD27
+ More
A0A194QYK5 A0A154PRA4 A0A0P6EJQ9 A0A0P5M057 A0A0N8E160 A0A0P6IQX4 A0A0P6AW20 A0A0P5T8N9 A0A0P5Y6Y2 A0A0P5YU19 K7J2X6 A0A232FEJ5 A0A2A3ECB7 E2B371 A0A088A7B4 A0A0C9R3K7 A0A310SU74 A0A0M8ZSJ0 A0A195E488 A0A026WZX8 A0A151X220 A0A195FIX4 F4W6G0 A0A158P272 A0A195AVJ2 K1QI76 T1JBR0 D1ZZV0 A0A2T7PMS1 A0A091HME1 J9LLJ4 A0A1W2WMP3 A0A091RW82 A0A1S3HRG7 V9K8J8 F6ZHD7 F6WUT8 A0A1S3K5E0 A0A091K569 A0A091QBN7 U3K106 A0A093GJL0 A0A099ZQ50 A0A091IV06 A0A0Q3U3C8 A0A1Y1ML03 W5PKH7 A0A3L8SK07 A0A091V691 A0A091LVX5 A0A094LA71
A0A194QYK5 A0A154PRA4 A0A0P6EJQ9 A0A0P5M057 A0A0N8E160 A0A0P6IQX4 A0A0P6AW20 A0A0P5T8N9 A0A0P5Y6Y2 A0A0P5YU19 K7J2X6 A0A232FEJ5 A0A2A3ECB7 E2B371 A0A088A7B4 A0A0C9R3K7 A0A310SU74 A0A0M8ZSJ0 A0A195E488 A0A026WZX8 A0A151X220 A0A195FIX4 F4W6G0 A0A158P272 A0A195AVJ2 K1QI76 T1JBR0 D1ZZV0 A0A2T7PMS1 A0A091HME1 J9LLJ4 A0A1W2WMP3 A0A091RW82 A0A1S3HRG7 V9K8J8 F6ZHD7 F6WUT8 A0A1S3K5E0 A0A091K569 A0A091QBN7 U3K106 A0A093GJL0 A0A099ZQ50 A0A091IV06 A0A0Q3U3C8 A0A1Y1ML03 W5PKH7 A0A3L8SK07 A0A091V691 A0A091LVX5 A0A094LA71
PDB
5THA
E-value=1.75655e-25,
Score=290
Ontologies
GO
PANTHER
Topology
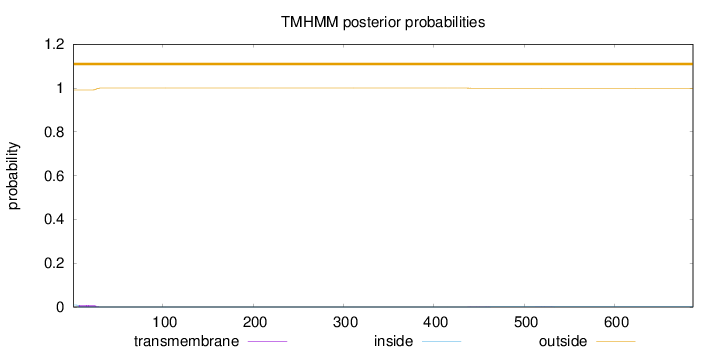
Length:
687
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.20099
Exp number, first 60 AAs:
0.18226
Total prob of N-in:
0.00914
outside
1 - 687
Population Genetic Test Statistics
Pi
201.296584
Theta
171.562104
Tajima's D
0.364613
CLR
0.204721
CSRT
0.475476226188691
Interpretation
Uncertain
