Gene
KWMTBOMO07663
Pre Gene Modal
BGIBMGA011097
Annotation
PREDICTED:_nose_resistant_to_fluoxetine_protein_6-like_[Papilio_polytes]
Location in the cell
PlasmaMembrane Reliability : 4.98
Sequence
CDS
ATGACCATTCTAACACTTAGCACCATTTTCGAAATCTTCATCATCTACATGGGGAAGAAAAAGGATTCCGTGATCACGGAACTGATAACGTCATTCTCTGTCATCAATAATGTAAAGAAGATTATATCAACGAAACAGCAGGGAAATCTTGGCTTAGAATGCGTAACTGGGATCAAAGCGCTGTCTATGATCTTTATTTTGGGCGGCCATGCCTGTCTATTCATAGCCAGTGGACCTGTTATGGATGCTACTGCTTGGGATAGACTAATCAGAACACCGCTAAATGGTTTCATGTTGAACAACGCTTTGCTCGTGGACACATTCTTGTTCCTGAGCGCTTTTCTCTTTTGCCGACTTCTTCTCATAGAACTGGATAAAAGACGAGGAAAACTTAACGTTATTCCAATTTTGGTATTCCGGTATATCAGGGTAACTCCGGCATATGCAGTTATCATATTATTCTATATGACTTGGTTCCCGAAGATCGGAGAGGGCCCGCTATGGGATGATCGACTGCTACTAGAGCGTGAGCGCTGCATGGAATCCTGGTGGGCTAATATCTTATACATCAATAACTACGTGAACACTGACCAAATGTGCATGTTCCAATCATGGTATTTATCTGTGGACACACAGTTATTCTTTGTGGCTCCGATATTCATCTACAGCCTGTGGCGTTGGAGGAAGTTTGGTACTTTTCTTCTGTCATTCGCTATTTTGGTGTCTATGTTGGTTCCATCTGTCATCACGTACAAAGACAAATTGGATCCAACGTTATTGTTTTTTGCCAAAGAATTCACGGATATTGTGTCAAACTTTTACTTTAAAGAGGCTTACATAAAGACGCACATGAAGATGATAACATATTGTATGGGGCTGTTGACCGGCTATGTACTACATCGTATACAGTCGACCAACTATCAAATGTCTTATCTCCTCAAGAGCTTTGCGTGGCTGACCAGCATTATCTTGGGAACTGTGACCACTTTTTCGGTCACCTTGTTTTATCAAGAATGGTACGAATATAGTAACATCGAGGCAGCTGCATACATTTCCCTGCACAAACTTGCCTGGGGCATCGCTAACGGGTGGCTGATCATTGCGTGTTCCACTGGCAATGGAGGTATCCTTGGGAGACTACTATCCTGGAAGTTCCTGGGTCCTGTAGCAAGATTGACATTCTGCGCATATTTAGTTAATGGAATTGTAGAGTTATACTACGTTGGACAGCTGAGGCATCCGATGCATATAACATTCTTCACTGTGGTCGCCTCGGCAACATCGCATATAGTACTGACGTTCTTCTTAGCCCTGCTTCTGTGTTTGACATTCGAATCACCGTTGCACGGTATTGAAAAGATTCTCCTAAAGAGATTTGCTCAACCGAAACATCGAGAAAATCCAGATTCAGTACAAACTTCAAGAAACACTAGTCAGTCCAGACTGGAGTCATAG
Protein
MTILTLSTIFEIFIIYMGKKKDSVITELITSFSVINNVKKIISTKQQGNLGLECVTGIKALSMIFILGGHACLFIASGPVMDATAWDRLIRTPLNGFMLNNALLVDTFLFLSAFLFCRLLLIELDKRRGKLNVIPILVFRYIRVTPAYAVIILFYMTWFPKIGEGPLWDDRLLLERERCMESWWANILYINNYVNTDQMCMFQSWYLSVDTQLFFVAPIFIYSLWRWRKFGTFLLSFAILVSMLVPSVITYKDKLDPTLLFFAKEFTDIVSNFYFKEAYIKTHMKMITYCMGLLTGYVLHRIQSTNYQMSYLLKSFAWLTSIILGTVTTFSVTLFYQEWYEYSNIEAAAYISLHKLAWGIANGWLIIACSTGNGGILGRLLSWKFLGPVARLTFCAYLVNGIVELYYVGQLRHPMHITFFTVVASATSHIVLTFFLALLLCLTFESPLHGIEKILLKRFAQPKHRENPDSVQTSRNTSQSRLES
Summary
Uniprot
A0A2A4J2E4
A0A2H1WFD8
A0A194PEK6
A0A212FDN9
A0A3S2LST9
W5JFI4
+ More
A0A034V3C2 A0A0K8UHZ0 A0A0K8VD85 W8BM55 A0A182SXU3 A0A182W4K6 A0A1J1HH25 A0A232EIQ2 K7J4D4 A0A182Y7U1 A0A182M972 A0A1A9VVB4 A0A182K8B2 A0A1A9Y8R3 A0A1B0BIV0 A0A1A9X1E4 A0A182R9I6 A0A1B0FMU0 A0A0C9PYJ9 A0A0T6B0W3 A0A182IAC3 A0NCC8 A0A182NDQ2 A0A182XEF8 A0A1B0D242 A0A182U2H9 A0A182V747 A0A182QX25 A0A0L0BZ72 A0A139W8X9 A0A182PNQ7 A0A023ESA0 A0A1A9Z0F3 A0A182FDU4 A0A182JM14 A0A1I8Q0V0 B0XH38 T1PHD7 A0A1I8M8L5 Q17BV5 E0VHW5 Q16E87 A0A182L859 E2B2T8 J9KT87 A0A1W4X317 A0A2S2Q8L1 D2A191 A0A2P8ZFS5 A0A2J7PXF5 A0A2J7PXE9 A0A1B6CQA8 A0A2J7PXG3 E0VHR7 A0A0C9R472 A0A2J7PZF1 A0A232EPY9 A0A087SXH8 A0A2J7QG24 A0A2J7PZI3 A0A1B6DQ61 K7ITV7 F4WNL2 E9G4F1 A0A151WGQ7 A0A1B6M9V1 A0A151IM87 A0A158NUM2 A0A210QXD8 A0A195EBZ0 A0A0L7R616 A0A0M9A7Z3 A0A195B8C9 A0A210Q427 A0A2P8XRP1 A0A3B0J7E0 B5DNQ9 A0A226EM11 E0VWD2 A0A2J7PZG8 A0A088A450 B4NE51 A0A0P4YQ91 A0A1A9Y8N8 A0A0P5YEI1 A0A1B6KJC8 A0A0M4EYZ3
A0A034V3C2 A0A0K8UHZ0 A0A0K8VD85 W8BM55 A0A182SXU3 A0A182W4K6 A0A1J1HH25 A0A232EIQ2 K7J4D4 A0A182Y7U1 A0A182M972 A0A1A9VVB4 A0A182K8B2 A0A1A9Y8R3 A0A1B0BIV0 A0A1A9X1E4 A0A182R9I6 A0A1B0FMU0 A0A0C9PYJ9 A0A0T6B0W3 A0A182IAC3 A0NCC8 A0A182NDQ2 A0A182XEF8 A0A1B0D242 A0A182U2H9 A0A182V747 A0A182QX25 A0A0L0BZ72 A0A139W8X9 A0A182PNQ7 A0A023ESA0 A0A1A9Z0F3 A0A182FDU4 A0A182JM14 A0A1I8Q0V0 B0XH38 T1PHD7 A0A1I8M8L5 Q17BV5 E0VHW5 Q16E87 A0A182L859 E2B2T8 J9KT87 A0A1W4X317 A0A2S2Q8L1 D2A191 A0A2P8ZFS5 A0A2J7PXF5 A0A2J7PXE9 A0A1B6CQA8 A0A2J7PXG3 E0VHR7 A0A0C9R472 A0A2J7PZF1 A0A232EPY9 A0A087SXH8 A0A2J7QG24 A0A2J7PZI3 A0A1B6DQ61 K7ITV7 F4WNL2 E9G4F1 A0A151WGQ7 A0A1B6M9V1 A0A151IM87 A0A158NUM2 A0A210QXD8 A0A195EBZ0 A0A0L7R616 A0A0M9A7Z3 A0A195B8C9 A0A210Q427 A0A2P8XRP1 A0A3B0J7E0 B5DNQ9 A0A226EM11 E0VWD2 A0A2J7PZG8 A0A088A450 B4NE51 A0A0P4YQ91 A0A1A9Y8N8 A0A0P5YEI1 A0A1B6KJC8 A0A0M4EYZ3
Pubmed
EMBL
NWSH01003464
PCG66235.1
ODYU01008317
SOQ51800.1
KQ459606
KPI91124.1
+ More
AGBW02009017 OWR51851.1 RSAL01000275 RVE43113.1 ADMH02001642 ETN61640.1 GAKP01021925 GAKP01021923 JAC37029.1 GDHF01026027 JAI26287.1 GDHF01015522 JAI36792.1 GAMC01012119 JAB94436.1 CVRI01000004 CRK87301.1 NNAY01004216 OXU18191.1 AXCM01008444 JXJN01015181 CCAG010022781 GBYB01006603 JAG76370.1 LJIG01016315 KRT80969.1 APCN01001143 AAAB01008846 EAU77249.3 AJVK01002698 AJVK01002699 AJVK01002700 AXCN02001261 JRES01001125 KNC25328.1 KQ972879 KXZ75738.1 GAPW01001480 JAC12118.1 DS233096 EDS28047.1 KA648181 AFP62810.1 CH477316 EAT43757.1 DS235172 EEB12971.1 CH478951 EAT32544.1 GL445241 EFN89991.1 ABLF02022069 GGMS01004866 MBY74069.1 KQ971338 EFA01566.2 PYGN01000071 PSN55353.1 NEVH01020859 PNF21017.1 PNF21015.1 GEDC01021708 JAS15590.1 PNF21016.1 DS235171 EEB12840.1 GBYB01007612 JAG77379.1 NEVH01020335 PNF21711.1 NNAY01002866 OXU20424.1 KK112404 KFM57567.1 NEVH01014836 PNF27544.1 PNF21736.1 GEDC01009491 JAS27807.1 GL888237 EGI64205.1 GL732532 EFX85286.1 KQ983150 KYQ47033.1 GEBQ01007264 JAT32713.1 KQ977063 KYN05983.1 ADTU01002856 NEDP02001363 OWF53373.1 KQ979074 KYN22745.1 KQ414648 KOC66332.1 KQ435728 KOX77773.1 KQ976565 KYM80500.1 NEDP02005089 OWF43498.1 PYGN01001466 PSN34669.1 OUUW01000003 SPP77997.1 CH379065 EDY73161.1 LNIX01000003 OXA58187.1 DS235819 EEB17717.1 PNF21712.1 CH964239 EDW82020.1 GDIP01226213 JAI97188.1 GDIP01058848 JAM44867.1 GEBQ01028428 JAT11549.1 CP012528 ALC49280.1
AGBW02009017 OWR51851.1 RSAL01000275 RVE43113.1 ADMH02001642 ETN61640.1 GAKP01021925 GAKP01021923 JAC37029.1 GDHF01026027 JAI26287.1 GDHF01015522 JAI36792.1 GAMC01012119 JAB94436.1 CVRI01000004 CRK87301.1 NNAY01004216 OXU18191.1 AXCM01008444 JXJN01015181 CCAG010022781 GBYB01006603 JAG76370.1 LJIG01016315 KRT80969.1 APCN01001143 AAAB01008846 EAU77249.3 AJVK01002698 AJVK01002699 AJVK01002700 AXCN02001261 JRES01001125 KNC25328.1 KQ972879 KXZ75738.1 GAPW01001480 JAC12118.1 DS233096 EDS28047.1 KA648181 AFP62810.1 CH477316 EAT43757.1 DS235172 EEB12971.1 CH478951 EAT32544.1 GL445241 EFN89991.1 ABLF02022069 GGMS01004866 MBY74069.1 KQ971338 EFA01566.2 PYGN01000071 PSN55353.1 NEVH01020859 PNF21017.1 PNF21015.1 GEDC01021708 JAS15590.1 PNF21016.1 DS235171 EEB12840.1 GBYB01007612 JAG77379.1 NEVH01020335 PNF21711.1 NNAY01002866 OXU20424.1 KK112404 KFM57567.1 NEVH01014836 PNF27544.1 PNF21736.1 GEDC01009491 JAS27807.1 GL888237 EGI64205.1 GL732532 EFX85286.1 KQ983150 KYQ47033.1 GEBQ01007264 JAT32713.1 KQ977063 KYN05983.1 ADTU01002856 NEDP02001363 OWF53373.1 KQ979074 KYN22745.1 KQ414648 KOC66332.1 KQ435728 KOX77773.1 KQ976565 KYM80500.1 NEDP02005089 OWF43498.1 PYGN01001466 PSN34669.1 OUUW01000003 SPP77997.1 CH379065 EDY73161.1 LNIX01000003 OXA58187.1 DS235819 EEB17717.1 PNF21712.1 CH964239 EDW82020.1 GDIP01226213 JAI97188.1 GDIP01058848 JAM44867.1 GEBQ01028428 JAT11549.1 CP012528 ALC49280.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000283053
UP000000673
UP000075901
+ More
UP000075920 UP000183832 UP000215335 UP000002358 UP000076408 UP000075883 UP000078200 UP000075881 UP000092443 UP000092460 UP000091820 UP000075900 UP000092444 UP000075840 UP000007062 UP000075884 UP000076407 UP000092462 UP000075902 UP000075903 UP000075886 UP000037069 UP000007266 UP000075885 UP000092445 UP000069272 UP000075880 UP000095300 UP000002320 UP000095301 UP000008820 UP000009046 UP000075882 UP000008237 UP000007819 UP000192223 UP000245037 UP000235965 UP000054359 UP000007755 UP000000305 UP000075809 UP000078542 UP000005205 UP000242188 UP000078492 UP000053825 UP000053105 UP000078540 UP000268350 UP000001819 UP000198287 UP000005203 UP000007798 UP000092553
UP000075920 UP000183832 UP000215335 UP000002358 UP000076408 UP000075883 UP000078200 UP000075881 UP000092443 UP000092460 UP000091820 UP000075900 UP000092444 UP000075840 UP000007062 UP000075884 UP000076407 UP000092462 UP000075902 UP000075903 UP000075886 UP000037069 UP000007266 UP000075885 UP000092445 UP000069272 UP000075880 UP000095300 UP000002320 UP000095301 UP000008820 UP000009046 UP000075882 UP000008237 UP000007819 UP000192223 UP000245037 UP000235965 UP000054359 UP000007755 UP000000305 UP000075809 UP000078542 UP000005205 UP000242188 UP000078492 UP000053825 UP000053105 UP000078540 UP000268350 UP000001819 UP000198287 UP000005203 UP000007798 UP000092553
Interpro
ProteinModelPortal
A0A2A4J2E4
A0A2H1WFD8
A0A194PEK6
A0A212FDN9
A0A3S2LST9
W5JFI4
+ More
A0A034V3C2 A0A0K8UHZ0 A0A0K8VD85 W8BM55 A0A182SXU3 A0A182W4K6 A0A1J1HH25 A0A232EIQ2 K7J4D4 A0A182Y7U1 A0A182M972 A0A1A9VVB4 A0A182K8B2 A0A1A9Y8R3 A0A1B0BIV0 A0A1A9X1E4 A0A182R9I6 A0A1B0FMU0 A0A0C9PYJ9 A0A0T6B0W3 A0A182IAC3 A0NCC8 A0A182NDQ2 A0A182XEF8 A0A1B0D242 A0A182U2H9 A0A182V747 A0A182QX25 A0A0L0BZ72 A0A139W8X9 A0A182PNQ7 A0A023ESA0 A0A1A9Z0F3 A0A182FDU4 A0A182JM14 A0A1I8Q0V0 B0XH38 T1PHD7 A0A1I8M8L5 Q17BV5 E0VHW5 Q16E87 A0A182L859 E2B2T8 J9KT87 A0A1W4X317 A0A2S2Q8L1 D2A191 A0A2P8ZFS5 A0A2J7PXF5 A0A2J7PXE9 A0A1B6CQA8 A0A2J7PXG3 E0VHR7 A0A0C9R472 A0A2J7PZF1 A0A232EPY9 A0A087SXH8 A0A2J7QG24 A0A2J7PZI3 A0A1B6DQ61 K7ITV7 F4WNL2 E9G4F1 A0A151WGQ7 A0A1B6M9V1 A0A151IM87 A0A158NUM2 A0A210QXD8 A0A195EBZ0 A0A0L7R616 A0A0M9A7Z3 A0A195B8C9 A0A210Q427 A0A2P8XRP1 A0A3B0J7E0 B5DNQ9 A0A226EM11 E0VWD2 A0A2J7PZG8 A0A088A450 B4NE51 A0A0P4YQ91 A0A1A9Y8N8 A0A0P5YEI1 A0A1B6KJC8 A0A0M4EYZ3
A0A034V3C2 A0A0K8UHZ0 A0A0K8VD85 W8BM55 A0A182SXU3 A0A182W4K6 A0A1J1HH25 A0A232EIQ2 K7J4D4 A0A182Y7U1 A0A182M972 A0A1A9VVB4 A0A182K8B2 A0A1A9Y8R3 A0A1B0BIV0 A0A1A9X1E4 A0A182R9I6 A0A1B0FMU0 A0A0C9PYJ9 A0A0T6B0W3 A0A182IAC3 A0NCC8 A0A182NDQ2 A0A182XEF8 A0A1B0D242 A0A182U2H9 A0A182V747 A0A182QX25 A0A0L0BZ72 A0A139W8X9 A0A182PNQ7 A0A023ESA0 A0A1A9Z0F3 A0A182FDU4 A0A182JM14 A0A1I8Q0V0 B0XH38 T1PHD7 A0A1I8M8L5 Q17BV5 E0VHW5 Q16E87 A0A182L859 E2B2T8 J9KT87 A0A1W4X317 A0A2S2Q8L1 D2A191 A0A2P8ZFS5 A0A2J7PXF5 A0A2J7PXE9 A0A1B6CQA8 A0A2J7PXG3 E0VHR7 A0A0C9R472 A0A2J7PZF1 A0A232EPY9 A0A087SXH8 A0A2J7QG24 A0A2J7PZI3 A0A1B6DQ61 K7ITV7 F4WNL2 E9G4F1 A0A151WGQ7 A0A1B6M9V1 A0A151IM87 A0A158NUM2 A0A210QXD8 A0A195EBZ0 A0A0L7R616 A0A0M9A7Z3 A0A195B8C9 A0A210Q427 A0A2P8XRP1 A0A3B0J7E0 B5DNQ9 A0A226EM11 E0VWD2 A0A2J7PZG8 A0A088A450 B4NE51 A0A0P4YQ91 A0A1A9Y8N8 A0A0P5YEI1 A0A1B6KJC8 A0A0M4EYZ3
Ontologies
GO
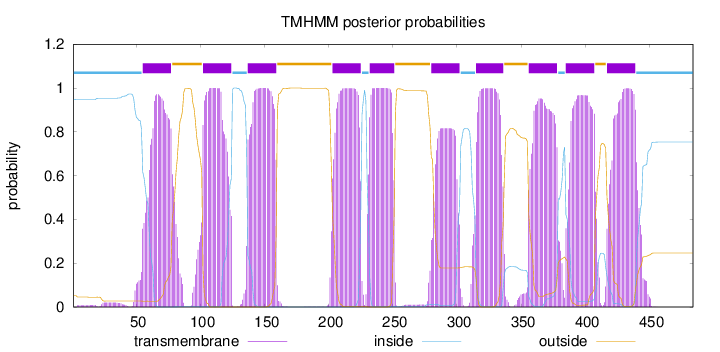
Topology
Length:
484
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
212.64135
Exp number, first 60 AAs:
3.97796
Total prob of N-in:
0.94782
inside
1 - 54
TMhelix
55 - 77
outside
78 - 101
TMhelix
102 - 124
inside
125 - 136
TMhelix
137 - 159
outside
160 - 202
TMhelix
203 - 225
inside
226 - 231
TMhelix
232 - 251
outside
252 - 279
TMhelix
280 - 302
inside
303 - 314
TMhelix
315 - 336
outside
337 - 355
TMhelix
356 - 378
inside
379 - 384
TMhelix
385 - 407
outside
408 - 416
TMhelix
417 - 439
inside
440 - 484
Population Genetic Test Statistics
Pi
159.426329
Theta
193.276771
Tajima's D
-1.125308
CLR
10.380409
CSRT
0.115894205289736
Interpretation
Uncertain
