Gene
KWMTBOMO07661
Pre Gene Modal
BGIBMGA001288
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_nose_resistant_to_fluoxetine_protein_6-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.834
Sequence
CDS
ATGGCGTCGCGCGCATGCGGTTTGATACTCACACTGTTCACCAGCGCGGTACTGTCGCGGGACTGCATCGAACTAACCATCAGCGACAAAAGAATCATAACAGAAACAATTCTAAATAGTACCATATTAAAGTTCCAACACGAGAATGATACGGTCGTTGAGAGTTCCCGGTTTAATATTGTGCAGTTGTGGGAGAGATTCCCGAAGGTTGTTGTGCCTAGGAACGGTACTAGTGAACAGTGCAGGAGAGACAGCCAGCTGTATATGGACAGTTTGGACCGACTGGAGTTATGGGCTTTGAAAATGTTTGACGCAACGGCCAAGCCCCCGTCTGGCATACTGAGCGGCAATGGAAACCAATATGGCGACTTTGACGAATGTCTGAGCATCGACGCCGCTGTCCGAGGGAAGTACTGCCTTGCCTCACTCATCATCAACTTCGGGGATGAGCAGAAATATAAAAAATTGGACTATCTCATTCACAGCGGACATTATATCAGGAGTAATATTACAGATATGGGTCACAGAGTTCCGCGGTATTCCAGTCTGCTTTGGGGCATCTGCATCCCAAGCTCGTGCAGTAGCTTCGACCTGGAAGAGGAGCTGTCGTACCGTCTATCACATCTCGGCATCACAGCCAAAGTACACAACACTATGTGCACAGTGAAGAATGTTAAACGTCCTTATTCGTTTGGGAAAGCCTTGTCCATATCTTTCTTCTTAGCGATACTGCTACTGTTAAGTCTTGGAACTATTTTGGACGACGACAAAGATGGCGTGACAAATTTCGAGAAACTACTTAACGCTTTTTCGTTGAAGCGTAATCTGAGGAAAGTGTTTGATCTGTCCGACGCAGCGACTAGGGTCAAAGGAGTGGACGCATTCAGGGGTATCAATGCCCTTGCTTTGCTCGTTGCGCATAAGTCAATGGCCATGGCGCATAACCCTTACGTTAACAAAGTTAGCTTCTCTGAGGTATTCGGAATGCCGTGGGCTGTGATCGGTCGCTCAGCGATACTATACACAGACAGTTTCCTGTACCTCAGTGGGTTCCTGAACGCTTACAACCTCCTGCAAGAGCTCGAGAGGAAAGGCACGATCGACTTGAAGAGCAGGCTCATCAACCGATGGTTTAGGTTATTTCCGTTGTTCGTGAGCCTGATGTTGTTCTGCACGTACATCCTGCCTGACATCAGCAACGGACCCCAGTGGAACCTCGTCGTGGAAGAGCACAGCAGAGTCTGCGAGAAGAACATGTGGAAGAGCTTCCTTTTCATTCACAACTATTTTGGATTCGAGGAAATGTGTCTGACGCACACGCATCAGATCGGTATGGACATGCAGCTGTACGTAGCGACCTTGCCTCTGATGCTTCTGTTCTGGAAAGCCAGGACCATTGGCTGGACCGTCTTCGCCTCCATCGCCGTCGGCTCCACTGTACTCAGATATCTTGCCATCCACTGGTACGACATCAGCATGTTCGTCTACTACGGAATATCAGTCCAGAAGTTACTGGACGCAGCAAGATATTCTTATATTCTGCCGACTCACAGAGCAACCATTTACCTAATCGGAGTAGCGATGGCCTACGTCATTAGAACGAAGAAATTGAACTTTACTCTCTCAAATAGCCAAGTAAGACTATTCTGGGCTTCGTGTTTCGCGTTAGCCGCCTTCACCATAGTGCACCCGTACTGGATGGGTCTGGAGGGGTACCAGTACGAGCACGGACCCGCCGCTCTGTTCGCGGCCCTGTCCCCTATCATGTGGGGCTTGTTCATGTGCATGATGCACTGGGCCGTGTGTAATGATTACGCTGGTTCTGGCACCGCCTTCCTGGAGTCGAGAGTGTTCAAATTCTTCAACAAAATCGCGTACAGTGTATACCTGACCCAGTTCCCGATCTTCTTCTTCACGGTCGGAGTCCAGAGGCACGCGGAATACTACTCGCCATTACTTCTGTTCCACATACCGGAGATGTTCGCAATCTTTGCGATATCCATAATGACGACAGTGGCCATCGAAATGCCCTTCAATCAAGTTTACAGGATATATTTCGGGAAATCACAGACGAAATTAAAAGAAAATTGA
Protein
MASRACGLILTLFTSAVLSRDCIELTISDKRIITETILNSTILKFQHENDTVVESSRFNIVQLWERFPKVVVPRNGTSEQCRRDSQLYMDSLDRLELWALKMFDATAKPPSGILSGNGNQYGDFDECLSIDAAVRGKYCLASLIINFGDEQKYKKLDYLIHSGHYIRSNITDMGHRVPRYSSLLWGICIPSSCSSFDLEEELSYRLSHLGITAKVHNTMCTVKNVKRPYSFGKALSISFFLAILLLLSLGTILDDDKDGVTNFEKLLNAFSLKRNLRKVFDLSDAATRVKGVDAFRGINALALLVAHKSMAMAHNPYVNKVSFSEVFGMPWAVIGRSAILYTDSFLYLSGFLNAYNLLQELERKGTIDLKSRLINRWFRLFPLFVSLMLFCTYILPDISNGPQWNLVVEEHSRVCEKNMWKSFLFIHNYFGFEEMCLTHTHQIGMDMQLYVATLPLMLLFWKARTIGWTVFASIAVGSTVLRYLAIHWYDISMFVYYGISVQKLLDAARYSYILPTHRATIYLIGVAMAYVIRTKKLNFTLSNSQVRLFWASCFALAAFTIVHPYWMGLEGYQYEHGPAALFAALSPIMWGLFMCMMHWAVCNDYAGSGTAFLESRVFKFFNKIAYSVYLTQFPIFFFTVGVQRHAEYYSPLLLFHIPEMFAIFAISIMTTVAIEMPFNQVYRIYFGKSQTKLKEN
Summary
Uniprot
A0A2A4J5M9
S4NN86
A0A3S2L120
A0A194PJ59
A0A2H1V3D5
A0A0L7L540
+ More
A0A067RNE3 A0A2J7PZG9 A0A2J7PZH2 A0A151J6N9 A0A2H8TWX1 A0A195CEC0 A0A0C9R9E4 A0A195F9R0 A0A151WFM3 J9K276 A0A195BX19 A0A1B6FL70 A0A232F722 K7IVS5 A0A2A3E408 E2B332 E0VHD6 E2A1E7 A0A146LAZ8 D6WLW1 A0A026W289 A0A3L8E2C0 A0A1W4X0F9 A0A0M9A7Q6 A0A0L7QTP8 F4WZ08 A0A0A1X526 A0A1W4V3G6 B3NUU8 B4PWA9 A0A034VQ95 B4IJA1 A0A0K8UK28 W8BUT3 A0A0K8VRM6 Q8IR42 T1HRZ8 A0A1B6C2J6 B3MR66 A0A1A9W8H9 A0A0A1X8X1 A0A310SLF0 A0A2J7QEL1 A0A336K276 B4H3M0 A0A0L0C729 A0A3B0JF37 A0A1Y1NCM1 B4N225 A0A154PKX0 A0A1Q3FQY8 A0A1Q3FR05 A0A1Q3FR09 A0A0M5J3K9 A0A1Q3FQU3 A0A067R775 B0WSU1 B4JMK6 A0A1I8Q534 B4L5A9 A0A182KF62 B4NUG5 A0A1I8N9P9 Q17L73 A0A182XXW2 A0A182Q2U8 A7USK5 A0A182NAC4 B4MAA6 A0A182UJJ0 A0A182MDC1 A0A1S4G9H5 A0A182W5F1 A0A182F9X6 A0A2S2Q8T7 A0A3B0JDU8 A0A182VAJ4 A0A182PV09 A0A1B6LMI1 A0A182X7C7 U4UBI8 A0A240PJW8 W5JLB6 A0A2P8ZED5 A0A084VQ88 A0A336N027 E0VHW8 A0A1B0CZ93
A0A067RNE3 A0A2J7PZG9 A0A2J7PZH2 A0A151J6N9 A0A2H8TWX1 A0A195CEC0 A0A0C9R9E4 A0A195F9R0 A0A151WFM3 J9K276 A0A195BX19 A0A1B6FL70 A0A232F722 K7IVS5 A0A2A3E408 E2B332 E0VHD6 E2A1E7 A0A146LAZ8 D6WLW1 A0A026W289 A0A3L8E2C0 A0A1W4X0F9 A0A0M9A7Q6 A0A0L7QTP8 F4WZ08 A0A0A1X526 A0A1W4V3G6 B3NUU8 B4PWA9 A0A034VQ95 B4IJA1 A0A0K8UK28 W8BUT3 A0A0K8VRM6 Q8IR42 T1HRZ8 A0A1B6C2J6 B3MR66 A0A1A9W8H9 A0A0A1X8X1 A0A310SLF0 A0A2J7QEL1 A0A336K276 B4H3M0 A0A0L0C729 A0A3B0JF37 A0A1Y1NCM1 B4N225 A0A154PKX0 A0A1Q3FQY8 A0A1Q3FR05 A0A1Q3FR09 A0A0M5J3K9 A0A1Q3FQU3 A0A067R775 B0WSU1 B4JMK6 A0A1I8Q534 B4L5A9 A0A182KF62 B4NUG5 A0A1I8N9P9 Q17L73 A0A182XXW2 A0A182Q2U8 A7USK5 A0A182NAC4 B4MAA6 A0A182UJJ0 A0A182MDC1 A0A1S4G9H5 A0A182W5F1 A0A182F9X6 A0A2S2Q8T7 A0A3B0JDU8 A0A182VAJ4 A0A182PV09 A0A1B6LMI1 A0A182X7C7 U4UBI8 A0A240PJW8 W5JLB6 A0A2P8ZED5 A0A084VQ88 A0A336N027 E0VHW8 A0A1B0CZ93
Pubmed
23622113
26354079
26227816
24845553
28648823
20075255
+ More
20798317 20566863 26823975 18362917 19820115 24508170 30249741 21719571 25830018 17994087 17550304 25348373 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 28004739 25315136 17510324 25244985 12364791 23537049 20920257 23761445 29403074 24438588
20798317 20566863 26823975 18362917 19820115 24508170 30249741 21719571 25830018 17994087 17550304 25348373 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 28004739 25315136 17510324 25244985 12364791 23537049 20920257 23761445 29403074 24438588
EMBL
NWSH01003083
PCG66978.1
GAIX01014041
JAA78519.1
RSAL01000275
RVE43115.1
+ More
KQ459606 KPI91120.1 ODYU01000488 SOQ35353.1 JTDY01002971 KOB70394.1 KK852566 KDR21234.1 NEVH01020335 PNF21733.1 PNF21734.1 KQ979863 KYN18772.1 GFXV01006990 MBW18795.1 KQ977873 KYM99147.1 GBYB01013028 GBYB01013029 JAG82795.1 JAG82796.1 KQ981727 KYN37116.1 KQ983211 KYQ46623.1 ABLF02017170 ABLF02028298 KQ976396 KYM92835.1 GECZ01018848 JAS50921.1 NNAY01000843 OXU26238.1 KZ288387 PBC26430.1 GL445285 EFN89881.1 DS235169 EEB12792.1 GL435766 EFN72620.1 GDHC01013884 JAQ04745.1 KQ971343 EFA04173.1 KK107468 EZA50205.1 QOIP01000001 RLU26672.1 KQ435719 KOX78937.1 KQ414740 KOC62037.1 GL888463 EGI60662.1 GBXI01007863 JAD06429.1 CH954180 EDV47046.1 CM000162 EDX01740.1 GAKP01013466 JAC45486.1 CH480847 EDW51080.1 GDHF01025619 JAI26695.1 GAMC01013666 GAMC01013665 JAB92889.1 GDHF01010821 JAI41493.1 AE014298 BT021240 AAN09655.2 AAX33388.1 ACPB03019813 GEDC01029819 JAS07479.1 CH902622 EDV34271.1 GBXI01007102 JAD07190.1 KQ764286 OAD54557.1 NEVH01015308 PNF27022.1 UFQS01000048 UFQT01000048 SSW98567.1 SSX18953.1 CH479207 EDW30971.1 JRES01000926 KNC27239.1 OUUW01000003 SPP78822.1 GEZM01009745 JAV94345.1 CH963925 EDW78414.2 KQ434950 KZC12506.1 GFDL01005066 JAV29979.1 GFDL01005046 JAV29999.1 GFDL01005060 JAV29985.1 CP012528 ALC49761.1 GFDL01005088 JAV29957.1 KK852657 KDR19201.1 DS232076 EDS34051.1 CH916371 EDV91949.1 CH933811 EDW06368.2 CH983974 EDX16612.1 CH477218 EAT47422.1 AXCN02001441 AAAB01008847 EDO64293.2 CH940655 EDW66165.1 AXCM01000689 GGMS01004946 MBY74149.1 SPP78823.1 GEBQ01027792 GEBQ01015188 JAT12185.1 JAT24789.1 KB632279 ERL91294.1 ADMH02000751 ETN65172.1 PYGN01000081 PSN54851.1 ATLV01015148 KE525003 KFB40132.1 UFQT01003846 SSX35311.1 DS235172 EEB12974.1 AJVK01002152
KQ459606 KPI91120.1 ODYU01000488 SOQ35353.1 JTDY01002971 KOB70394.1 KK852566 KDR21234.1 NEVH01020335 PNF21733.1 PNF21734.1 KQ979863 KYN18772.1 GFXV01006990 MBW18795.1 KQ977873 KYM99147.1 GBYB01013028 GBYB01013029 JAG82795.1 JAG82796.1 KQ981727 KYN37116.1 KQ983211 KYQ46623.1 ABLF02017170 ABLF02028298 KQ976396 KYM92835.1 GECZ01018848 JAS50921.1 NNAY01000843 OXU26238.1 KZ288387 PBC26430.1 GL445285 EFN89881.1 DS235169 EEB12792.1 GL435766 EFN72620.1 GDHC01013884 JAQ04745.1 KQ971343 EFA04173.1 KK107468 EZA50205.1 QOIP01000001 RLU26672.1 KQ435719 KOX78937.1 KQ414740 KOC62037.1 GL888463 EGI60662.1 GBXI01007863 JAD06429.1 CH954180 EDV47046.1 CM000162 EDX01740.1 GAKP01013466 JAC45486.1 CH480847 EDW51080.1 GDHF01025619 JAI26695.1 GAMC01013666 GAMC01013665 JAB92889.1 GDHF01010821 JAI41493.1 AE014298 BT021240 AAN09655.2 AAX33388.1 ACPB03019813 GEDC01029819 JAS07479.1 CH902622 EDV34271.1 GBXI01007102 JAD07190.1 KQ764286 OAD54557.1 NEVH01015308 PNF27022.1 UFQS01000048 UFQT01000048 SSW98567.1 SSX18953.1 CH479207 EDW30971.1 JRES01000926 KNC27239.1 OUUW01000003 SPP78822.1 GEZM01009745 JAV94345.1 CH963925 EDW78414.2 KQ434950 KZC12506.1 GFDL01005066 JAV29979.1 GFDL01005046 JAV29999.1 GFDL01005060 JAV29985.1 CP012528 ALC49761.1 GFDL01005088 JAV29957.1 KK852657 KDR19201.1 DS232076 EDS34051.1 CH916371 EDV91949.1 CH933811 EDW06368.2 CH983974 EDX16612.1 CH477218 EAT47422.1 AXCN02001441 AAAB01008847 EDO64293.2 CH940655 EDW66165.1 AXCM01000689 GGMS01004946 MBY74149.1 SPP78823.1 GEBQ01027792 GEBQ01015188 JAT12185.1 JAT24789.1 KB632279 ERL91294.1 ADMH02000751 ETN65172.1 PYGN01000081 PSN54851.1 ATLV01015148 KE525003 KFB40132.1 UFQT01003846 SSX35311.1 DS235172 EEB12974.1 AJVK01002152
Proteomes
UP000218220
UP000283053
UP000053268
UP000037510
UP000027135
UP000235965
+ More
UP000078492 UP000078542 UP000078541 UP000075809 UP000007819 UP000078540 UP000215335 UP000002358 UP000242457 UP000008237 UP000009046 UP000000311 UP000007266 UP000053097 UP000279307 UP000192223 UP000053105 UP000053825 UP000007755 UP000192221 UP000008711 UP000002282 UP000001292 UP000000803 UP000015103 UP000007801 UP000091820 UP000008744 UP000037069 UP000268350 UP000007798 UP000076502 UP000092553 UP000002320 UP000001070 UP000095300 UP000009192 UP000075881 UP000000304 UP000095301 UP000008820 UP000076408 UP000075886 UP000007062 UP000075884 UP000008792 UP000075902 UP000075883 UP000075920 UP000069272 UP000075903 UP000075885 UP000076407 UP000030742 UP000075880 UP000000673 UP000245037 UP000030765 UP000092462
UP000078492 UP000078542 UP000078541 UP000075809 UP000007819 UP000078540 UP000215335 UP000002358 UP000242457 UP000008237 UP000009046 UP000000311 UP000007266 UP000053097 UP000279307 UP000192223 UP000053105 UP000053825 UP000007755 UP000192221 UP000008711 UP000002282 UP000001292 UP000000803 UP000015103 UP000007801 UP000091820 UP000008744 UP000037069 UP000268350 UP000007798 UP000076502 UP000092553 UP000002320 UP000001070 UP000095300 UP000009192 UP000075881 UP000000304 UP000095301 UP000008820 UP000076408 UP000075886 UP000007062 UP000075884 UP000008792 UP000075902 UP000075883 UP000075920 UP000069272 UP000075903 UP000075885 UP000076407 UP000030742 UP000075880 UP000000673 UP000245037 UP000030765 UP000092462
Pfam
PF01757 Acyl_transf_3
ProteinModelPortal
A0A2A4J5M9
S4NN86
A0A3S2L120
A0A194PJ59
A0A2H1V3D5
A0A0L7L540
+ More
A0A067RNE3 A0A2J7PZG9 A0A2J7PZH2 A0A151J6N9 A0A2H8TWX1 A0A195CEC0 A0A0C9R9E4 A0A195F9R0 A0A151WFM3 J9K276 A0A195BX19 A0A1B6FL70 A0A232F722 K7IVS5 A0A2A3E408 E2B332 E0VHD6 E2A1E7 A0A146LAZ8 D6WLW1 A0A026W289 A0A3L8E2C0 A0A1W4X0F9 A0A0M9A7Q6 A0A0L7QTP8 F4WZ08 A0A0A1X526 A0A1W4V3G6 B3NUU8 B4PWA9 A0A034VQ95 B4IJA1 A0A0K8UK28 W8BUT3 A0A0K8VRM6 Q8IR42 T1HRZ8 A0A1B6C2J6 B3MR66 A0A1A9W8H9 A0A0A1X8X1 A0A310SLF0 A0A2J7QEL1 A0A336K276 B4H3M0 A0A0L0C729 A0A3B0JF37 A0A1Y1NCM1 B4N225 A0A154PKX0 A0A1Q3FQY8 A0A1Q3FR05 A0A1Q3FR09 A0A0M5J3K9 A0A1Q3FQU3 A0A067R775 B0WSU1 B4JMK6 A0A1I8Q534 B4L5A9 A0A182KF62 B4NUG5 A0A1I8N9P9 Q17L73 A0A182XXW2 A0A182Q2U8 A7USK5 A0A182NAC4 B4MAA6 A0A182UJJ0 A0A182MDC1 A0A1S4G9H5 A0A182W5F1 A0A182F9X6 A0A2S2Q8T7 A0A3B0JDU8 A0A182VAJ4 A0A182PV09 A0A1B6LMI1 A0A182X7C7 U4UBI8 A0A240PJW8 W5JLB6 A0A2P8ZED5 A0A084VQ88 A0A336N027 E0VHW8 A0A1B0CZ93
A0A067RNE3 A0A2J7PZG9 A0A2J7PZH2 A0A151J6N9 A0A2H8TWX1 A0A195CEC0 A0A0C9R9E4 A0A195F9R0 A0A151WFM3 J9K276 A0A195BX19 A0A1B6FL70 A0A232F722 K7IVS5 A0A2A3E408 E2B332 E0VHD6 E2A1E7 A0A146LAZ8 D6WLW1 A0A026W289 A0A3L8E2C0 A0A1W4X0F9 A0A0M9A7Q6 A0A0L7QTP8 F4WZ08 A0A0A1X526 A0A1W4V3G6 B3NUU8 B4PWA9 A0A034VQ95 B4IJA1 A0A0K8UK28 W8BUT3 A0A0K8VRM6 Q8IR42 T1HRZ8 A0A1B6C2J6 B3MR66 A0A1A9W8H9 A0A0A1X8X1 A0A310SLF0 A0A2J7QEL1 A0A336K276 B4H3M0 A0A0L0C729 A0A3B0JF37 A0A1Y1NCM1 B4N225 A0A154PKX0 A0A1Q3FQY8 A0A1Q3FR05 A0A1Q3FR09 A0A0M5J3K9 A0A1Q3FQU3 A0A067R775 B0WSU1 B4JMK6 A0A1I8Q534 B4L5A9 A0A182KF62 B4NUG5 A0A1I8N9P9 Q17L73 A0A182XXW2 A0A182Q2U8 A7USK5 A0A182NAC4 B4MAA6 A0A182UJJ0 A0A182MDC1 A0A1S4G9H5 A0A182W5F1 A0A182F9X6 A0A2S2Q8T7 A0A3B0JDU8 A0A182VAJ4 A0A182PV09 A0A1B6LMI1 A0A182X7C7 U4UBI8 A0A240PJW8 W5JLB6 A0A2P8ZED5 A0A084VQ88 A0A336N027 E0VHW8 A0A1B0CZ93
Ontologies
GO
Topology
SignalP
Position: 1 - 19,
Likelihood: 0.922798
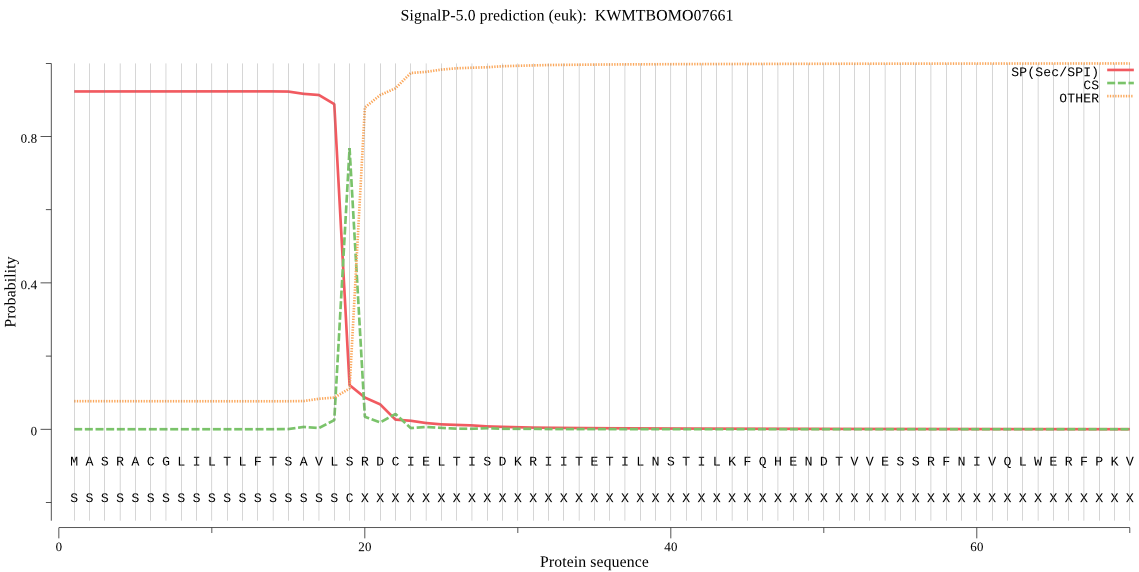
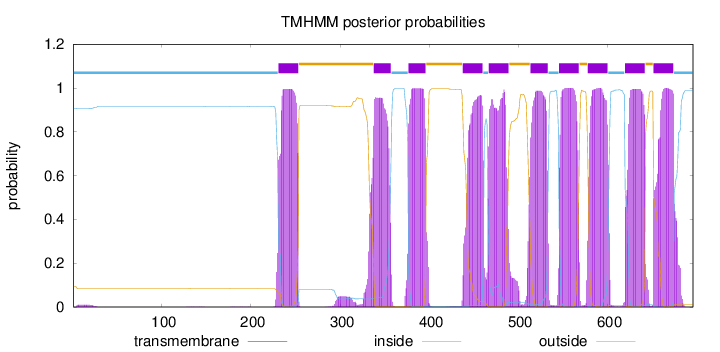
Length:
696
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
214.96245
Exp number, first 60 AAs:
0.19081
Total prob of N-in:
0.90621
inside
1 - 230
TMhelix
231 - 253
outside
254 - 337
TMhelix
338 - 357
inside
358 - 376
TMhelix
377 - 396
outside
397 - 437
TMhelix
438 - 460
inside
461 - 466
TMhelix
467 - 489
outside
490 - 513
TMhelix
514 - 533
inside
534 - 545
TMhelix
546 - 568
outside
569 - 577
TMhelix
578 - 600
inside
601 - 619
TMhelix
620 - 642
outside
643 - 651
TMhelix
652 - 674
inside
675 - 696
Population Genetic Test Statistics
Pi
204.728776
Theta
161.503492
Tajima's D
1.028294
CLR
0.344324
CSRT
0.663766811659417
Interpretation
Uncertain
