Gene
KWMTBOMO07660
Pre Gene Modal
BGIBMGA000856
Annotation
PREDICTED:_uncharacterized_protein_LOC106134578_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 4.216
Sequence
CDS
ATGGCGACCTGGATTTGGTTGGCGGCATTGTTCTGTGGGACGCACGCCGCAGTGCTACCACCACCGTGGGCGGACGTCAGAAGGAATCCCTGTGCCGCTCATCCGAGAGGCTGGCTTATGTTATACTGGCCGATCGATGGAAAATGCTATACAATTTATCAGAAAGGTCACCCGTGTCCCGAGACTCAAGAACTCAGTCCCGGTCGCTGGGGCGGAAGGACTATAGCAGAATGCAAGTGTCCGCCTGGGACGGCCCAGCTGCCGCATACAACGACATGCCACAAGCTATTCGAACGAGGTCCTTGCAGAGCCGGCGAGTACTTCGCGCCGGTCGAGGAGTCCTTCAACAAGCGAGGTGAACGTCAAGGTATGTGCGTGAGGCCGCAGCAGTGCGCTGACGAAACCTTGCTGTACTGGCCCGCTGATGGGAAGTGCTACTACAGGCTCAGTCAGGGCCCTTGTTACCCAGGTGCCATATTGGATATCGGAGACGATGGACTGGCGAACTGTACGTGTGATCCGGATAGTAGAAACTTCTGGCCGCTAGACGGAGGTTGCTATGCGCATTTCACCCGGGGACCCTGTGACAAGGGCCTGTTGTATCTGCCGGGACCAAGATGTGGCTGCGAGAGCCATATGCCGCATTATCACAACGAGACCGGGGGGTGTTATGAACTGGATTCCGTCGGCCCGTGTCCTCGAGGCCACACCTTCAGGATAACACAAGTGACAGCCCAAGGATCTAAAGCGGAGTGTAAATGTAAATCGTTCCACGCCAGAGCCGCTGACGGAGCTTGCTATAGACTCTACACCAGGGGACCTTGCGAACAGGATGAAATGATAGCGCGAGGGGGGCGATGTATCAAGGTTCCGTGTGCGAGAGGTCGTCTGTACGTCCCAGAGAGGCGTCGATGCTACCGGCCCGGCACGCCCGAGCCGTGTCCGGTCGGGGAGGTCGTCGCGTTCGACTTCGACGCCAGACCAGCGCTGGACGGGCTCAGTCATAACGGAGTTTGCGTCTGCGGCAGCGGGTCTTGTAATAGAAAAGAAGTCGAAGCGTGTACCGCCCGCAAAGGGACCGCTGCGTATGGCGGCGCTTGTCACATTCTCAACACTCAAGGTCCCTGCCCGGACGGATCCTGGCTCGTCAAGGAAGGAACCGGGAAAGTCAAGTGTCAATGTCGCCCTGGTCGCGTCCCTCCGGAGTGTAACGGAGACATCGACCCTCAAGCTAAGAGCTGA
Protein
MATWIWLAALFCGTHAAVLPPPWADVRRNPCAAHPRGWLMLYWPIDGKCYTIYQKGHPCPETQELSPGRWGGRTIAECKCPPGTAQLPHTTTCHKLFERGPCRAGEYFAPVEESFNKRGERQGMCVRPQQCADETLLYWPADGKCYYRLSQGPCYPGAILDIGDDGLANCTCDPDSRNFWPLDGGCYAHFTRGPCDKGLLYLPGPRCGCESHMPHYHNETGGCYELDSVGPCPRGHTFRITQVTAQGSKAECKCKSFHARAADGACYRLYTRGPCEQDEMIARGGRCIKVPCARGRLYVPERRRCYRPGTPEPCPVGEVVAFDFDARPALDGLSHNGVCVCGSGSCNRKEVEACTARKGTAAYGGACHILNTQGPCPDGSWLVKEGTGKVKCQCRPGRVPPECNGDIDPQAKS
Summary
Uniprot
H9IUC6
A0A3S2NBH7
A0A194PEK1
A0A212ESE5
A0A2A4J4A1
A0A0L7L4G5
+ More
A0A2H1WYL5 A0A194R3V9 A0A158NQE4 A0A151I2C8 A0A195CPZ9 A0A195EWF7 F4WGM4 A0A195DUU3 A0A3L8D8A9 A0A026WWC6 A0A088A251 E2B073 K7IW83 A0A310SE85 A0A0M9A4M3 A0A0L7R604 A0A154NW18 A0A232FLH1 A0A182NZX1 A0A182M2I8 A0A182W216 A0A182N301 A0A182JV54 A0A336L3Q8 Q176K7 Q7QH18 A0A336KCN1 A0A182RKE9 A0A182QAS2 B4KXM0 A0A182YBL8 A0A182HVX6 A0A182WUJ7 A0A0Q9XDX1 A0A182VJA6 B0WU93 A0A0L0CEA2 A0A182J5U6 A0A023EST2 A0A182FTP4 A0A0Q5UJE2 A0A1B0CZN5 B3NHN1 A0A0R1DXX5 B3M737 B4PIY0 A0A182U151 W5JC01 A0A1W4VMT7 A0A1W4VNI9 A0A1I8NB67 A0A0C9Q170 A0A1I8NB52 B4N526 A0A0Q9WJF3 B4LFL0 Q9VUL4 M9PFB1 B4J2X3 A0NAK3 A0A0J9RVE3 A0A0J9RVH0 A0A3B0JKX4 A0A3B0JY37 A0A3B0J7V4 A0A1I8Q9R6 A0A3B0JTI7 A0A3B0JQS4 A0A3B0J3P6 A0A1I8Q9U8 A0A2J7PZI8 A0A0J9RWR5 A0A0J9RVW0 Q2M199 A0A0R3P2W1 A0A151XH84 A0A0K8TVP3 A0A0K8WGH1 U4UNG3 W8C0N9 A0A034V4C9 A0A067RBW8 A0A1W4WQT9 N6TLR6 B4QKK3 B4GUE9 A0A0A1WTW3 A0A0A1XD75 A0A2A3EFG1 E0VBA6 E2BF07 D2A085 A0A0J7L195 E9IDI3
A0A2H1WYL5 A0A194R3V9 A0A158NQE4 A0A151I2C8 A0A195CPZ9 A0A195EWF7 F4WGM4 A0A195DUU3 A0A3L8D8A9 A0A026WWC6 A0A088A251 E2B073 K7IW83 A0A310SE85 A0A0M9A4M3 A0A0L7R604 A0A154NW18 A0A232FLH1 A0A182NZX1 A0A182M2I8 A0A182W216 A0A182N301 A0A182JV54 A0A336L3Q8 Q176K7 Q7QH18 A0A336KCN1 A0A182RKE9 A0A182QAS2 B4KXM0 A0A182YBL8 A0A182HVX6 A0A182WUJ7 A0A0Q9XDX1 A0A182VJA6 B0WU93 A0A0L0CEA2 A0A182J5U6 A0A023EST2 A0A182FTP4 A0A0Q5UJE2 A0A1B0CZN5 B3NHN1 A0A0R1DXX5 B3M737 B4PIY0 A0A182U151 W5JC01 A0A1W4VMT7 A0A1W4VNI9 A0A1I8NB67 A0A0C9Q170 A0A1I8NB52 B4N526 A0A0Q9WJF3 B4LFL0 Q9VUL4 M9PFB1 B4J2X3 A0NAK3 A0A0J9RVE3 A0A0J9RVH0 A0A3B0JKX4 A0A3B0JY37 A0A3B0J7V4 A0A1I8Q9R6 A0A3B0JTI7 A0A3B0JQS4 A0A3B0J3P6 A0A1I8Q9U8 A0A2J7PZI8 A0A0J9RWR5 A0A0J9RVW0 Q2M199 A0A0R3P2W1 A0A151XH84 A0A0K8TVP3 A0A0K8WGH1 U4UNG3 W8C0N9 A0A034V4C9 A0A067RBW8 A0A1W4WQT9 N6TLR6 B4QKK3 B4GUE9 A0A0A1WTW3 A0A0A1XD75 A0A2A3EFG1 E0VBA6 E2BF07 D2A085 A0A0J7L195 E9IDI3
Pubmed
19121390
26354079
22118469
26227816
21347285
21719571
+ More
30249741 24508170 20798317 20075255 28648823 17510324 12364791 14747013 17210077 17994087 18057021 25244985 26108605 24945155 17550304 20920257 23761445 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 9087549 22936249 15632085 23537049 24495485 25348373 24845553 25830018 20566863 18362917 19820115 21282665
30249741 24508170 20798317 20075255 28648823 17510324 12364791 14747013 17210077 17994087 18057021 25244985 26108605 24945155 17550304 20920257 23761445 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 9087549 22936249 15632085 23537049 24495485 25348373 24845553 25830018 20566863 18362917 19820115 21282665
EMBL
BABH01001342
RSAL01000275
RVE43116.1
KQ459606
KPI91119.1
AGBW02012839
+ More
OWR44361.1 NWSH01003083 PCG66977.1 JTDY01002971 KOB70393.1 ODYU01012017 SOQ58097.1 KQ460947 KPJ10526.1 ADTU01023159 KQ976539 KYM81306.1 KQ977444 KYN02705.1 KQ981948 KYN32568.1 GL888139 EGI66648.1 KQ980322 KYN16643.1 QOIP01000011 RLU16737.1 KK107078 EZA60380.1 GL444408 EFN60930.1 KQ762237 OAD55995.1 KQ435750 KOX76376.1 KQ414648 KOC66278.1 KQ434772 KZC03885.1 NNAY01000053 OXU31505.1 AXCM01000173 UFQS01001716 UFQT01001716 SSX11956.1 SSX31503.1 CH477386 EAT42074.1 AAAB01008823 EAA05517.3 UFQS01000062 UFQT01000062 SSW98820.1 SSX19206.1 AXCN02001144 CH933809 EDW19727.1 KRG06783.1 APCN01001435 KRG06784.1 DS232102 EDS34864.1 JRES01000502 KNC30580.1 GAPW01001256 JAC12342.1 CH954178 KQS43991.1 KQS43992.1 AJVK01009642 EDV51826.2 CM000159 KRK01931.1 CH902618 EDV40902.1 EDW94571.2 ADMH02001947 ETN60364.1 GBYB01007653 JAG77420.1 CH964101 EDW79465.1 CH940647 KRF84857.1 EDW70328.2 AE014296 AY051448 AAF49662.1 AAK92872.1 ADV37545.1 AGB94563.1 AHN58094.1 CH916366 EDV97143.1 AAAB01004400 EAU77984.2 CM002912 KMY99698.1 KMY99703.1 OUUW01000002 SPP76010.1 SPP76008.1 SPP76012.1 SPP76011.1 SPP76009.1 SPP76007.1 NEVH01020335 PNF21745.1 KMY99699.1 KMY99701.1 KMY99704.1 KMY99702.1 CH379069 EAL30676.3 KRT07522.1 KQ982156 KYQ59570.1 GDHF01033983 GDHF01005316 JAI18331.1 JAI46998.1 GDHF01023267 GDHF01019729 GDHF01002145 JAI29047.1 JAI32585.1 JAI50169.1 KB632375 ERL94008.1 GAMC01011366 GAMC01011365 GAMC01011364 JAB95189.1 GAKP01022347 JAC36605.1 KK852566 KDR21242.1 APGK01030817 KB740787 ENN78928.1 CM000363 EDX10514.1 CH479191 EDW26232.1 GBXI01011778 GBXI01009384 JAD02514.1 JAD04908.1 GBXI01004963 JAD09329.1 KZ288260 PBC30460.1 DS235023 EEB10661.1 GL447903 EFN85730.1 KQ971338 EFA01733.2 LBMM01001379 KMQ96416.1 GL762454 EFZ21415.1
OWR44361.1 NWSH01003083 PCG66977.1 JTDY01002971 KOB70393.1 ODYU01012017 SOQ58097.1 KQ460947 KPJ10526.1 ADTU01023159 KQ976539 KYM81306.1 KQ977444 KYN02705.1 KQ981948 KYN32568.1 GL888139 EGI66648.1 KQ980322 KYN16643.1 QOIP01000011 RLU16737.1 KK107078 EZA60380.1 GL444408 EFN60930.1 KQ762237 OAD55995.1 KQ435750 KOX76376.1 KQ414648 KOC66278.1 KQ434772 KZC03885.1 NNAY01000053 OXU31505.1 AXCM01000173 UFQS01001716 UFQT01001716 SSX11956.1 SSX31503.1 CH477386 EAT42074.1 AAAB01008823 EAA05517.3 UFQS01000062 UFQT01000062 SSW98820.1 SSX19206.1 AXCN02001144 CH933809 EDW19727.1 KRG06783.1 APCN01001435 KRG06784.1 DS232102 EDS34864.1 JRES01000502 KNC30580.1 GAPW01001256 JAC12342.1 CH954178 KQS43991.1 KQS43992.1 AJVK01009642 EDV51826.2 CM000159 KRK01931.1 CH902618 EDV40902.1 EDW94571.2 ADMH02001947 ETN60364.1 GBYB01007653 JAG77420.1 CH964101 EDW79465.1 CH940647 KRF84857.1 EDW70328.2 AE014296 AY051448 AAF49662.1 AAK92872.1 ADV37545.1 AGB94563.1 AHN58094.1 CH916366 EDV97143.1 AAAB01004400 EAU77984.2 CM002912 KMY99698.1 KMY99703.1 OUUW01000002 SPP76010.1 SPP76008.1 SPP76012.1 SPP76011.1 SPP76009.1 SPP76007.1 NEVH01020335 PNF21745.1 KMY99699.1 KMY99701.1 KMY99704.1 KMY99702.1 CH379069 EAL30676.3 KRT07522.1 KQ982156 KYQ59570.1 GDHF01033983 GDHF01005316 JAI18331.1 JAI46998.1 GDHF01023267 GDHF01019729 GDHF01002145 JAI29047.1 JAI32585.1 JAI50169.1 KB632375 ERL94008.1 GAMC01011366 GAMC01011365 GAMC01011364 JAB95189.1 GAKP01022347 JAC36605.1 KK852566 KDR21242.1 APGK01030817 KB740787 ENN78928.1 CM000363 EDX10514.1 CH479191 EDW26232.1 GBXI01011778 GBXI01009384 JAD02514.1 JAD04908.1 GBXI01004963 JAD09329.1 KZ288260 PBC30460.1 DS235023 EEB10661.1 GL447903 EFN85730.1 KQ971338 EFA01733.2 LBMM01001379 KMQ96416.1 GL762454 EFZ21415.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000007151
UP000218220
UP000037510
+ More
UP000053240 UP000005205 UP000078540 UP000078542 UP000078541 UP000007755 UP000078492 UP000279307 UP000053097 UP000005203 UP000000311 UP000002358 UP000053105 UP000053825 UP000076502 UP000215335 UP000075885 UP000075883 UP000075920 UP000075884 UP000075881 UP000008820 UP000007062 UP000075900 UP000075886 UP000009192 UP000076408 UP000075840 UP000076407 UP000075903 UP000002320 UP000037069 UP000075880 UP000069272 UP000008711 UP000092462 UP000002282 UP000007801 UP000075902 UP000000673 UP000192221 UP000095301 UP000007798 UP000008792 UP000000803 UP000001070 UP000268350 UP000095300 UP000235965 UP000001819 UP000075809 UP000030742 UP000027135 UP000192223 UP000019118 UP000000304 UP000008744 UP000242457 UP000009046 UP000008237 UP000007266 UP000036403
UP000053240 UP000005205 UP000078540 UP000078542 UP000078541 UP000007755 UP000078492 UP000279307 UP000053097 UP000005203 UP000000311 UP000002358 UP000053105 UP000053825 UP000076502 UP000215335 UP000075885 UP000075883 UP000075920 UP000075884 UP000075881 UP000008820 UP000007062 UP000075900 UP000075886 UP000009192 UP000076408 UP000075840 UP000076407 UP000075903 UP000002320 UP000037069 UP000075880 UP000069272 UP000008711 UP000092462 UP000002282 UP000007801 UP000075902 UP000000673 UP000192221 UP000095301 UP000007798 UP000008792 UP000000803 UP000001070 UP000268350 UP000095300 UP000235965 UP000001819 UP000075809 UP000030742 UP000027135 UP000192223 UP000019118 UP000000304 UP000008744 UP000242457 UP000009046 UP000008237 UP000007266 UP000036403
Pfam
PF16033 DUF4789
SUPFAM
SSF57184
SSF57184
ProteinModelPortal
H9IUC6
A0A3S2NBH7
A0A194PEK1
A0A212ESE5
A0A2A4J4A1
A0A0L7L4G5
+ More
A0A2H1WYL5 A0A194R3V9 A0A158NQE4 A0A151I2C8 A0A195CPZ9 A0A195EWF7 F4WGM4 A0A195DUU3 A0A3L8D8A9 A0A026WWC6 A0A088A251 E2B073 K7IW83 A0A310SE85 A0A0M9A4M3 A0A0L7R604 A0A154NW18 A0A232FLH1 A0A182NZX1 A0A182M2I8 A0A182W216 A0A182N301 A0A182JV54 A0A336L3Q8 Q176K7 Q7QH18 A0A336KCN1 A0A182RKE9 A0A182QAS2 B4KXM0 A0A182YBL8 A0A182HVX6 A0A182WUJ7 A0A0Q9XDX1 A0A182VJA6 B0WU93 A0A0L0CEA2 A0A182J5U6 A0A023EST2 A0A182FTP4 A0A0Q5UJE2 A0A1B0CZN5 B3NHN1 A0A0R1DXX5 B3M737 B4PIY0 A0A182U151 W5JC01 A0A1W4VMT7 A0A1W4VNI9 A0A1I8NB67 A0A0C9Q170 A0A1I8NB52 B4N526 A0A0Q9WJF3 B4LFL0 Q9VUL4 M9PFB1 B4J2X3 A0NAK3 A0A0J9RVE3 A0A0J9RVH0 A0A3B0JKX4 A0A3B0JY37 A0A3B0J7V4 A0A1I8Q9R6 A0A3B0JTI7 A0A3B0JQS4 A0A3B0J3P6 A0A1I8Q9U8 A0A2J7PZI8 A0A0J9RWR5 A0A0J9RVW0 Q2M199 A0A0R3P2W1 A0A151XH84 A0A0K8TVP3 A0A0K8WGH1 U4UNG3 W8C0N9 A0A034V4C9 A0A067RBW8 A0A1W4WQT9 N6TLR6 B4QKK3 B4GUE9 A0A0A1WTW3 A0A0A1XD75 A0A2A3EFG1 E0VBA6 E2BF07 D2A085 A0A0J7L195 E9IDI3
A0A2H1WYL5 A0A194R3V9 A0A158NQE4 A0A151I2C8 A0A195CPZ9 A0A195EWF7 F4WGM4 A0A195DUU3 A0A3L8D8A9 A0A026WWC6 A0A088A251 E2B073 K7IW83 A0A310SE85 A0A0M9A4M3 A0A0L7R604 A0A154NW18 A0A232FLH1 A0A182NZX1 A0A182M2I8 A0A182W216 A0A182N301 A0A182JV54 A0A336L3Q8 Q176K7 Q7QH18 A0A336KCN1 A0A182RKE9 A0A182QAS2 B4KXM0 A0A182YBL8 A0A182HVX6 A0A182WUJ7 A0A0Q9XDX1 A0A182VJA6 B0WU93 A0A0L0CEA2 A0A182J5U6 A0A023EST2 A0A182FTP4 A0A0Q5UJE2 A0A1B0CZN5 B3NHN1 A0A0R1DXX5 B3M737 B4PIY0 A0A182U151 W5JC01 A0A1W4VMT7 A0A1W4VNI9 A0A1I8NB67 A0A0C9Q170 A0A1I8NB52 B4N526 A0A0Q9WJF3 B4LFL0 Q9VUL4 M9PFB1 B4J2X3 A0NAK3 A0A0J9RVE3 A0A0J9RVH0 A0A3B0JKX4 A0A3B0JY37 A0A3B0J7V4 A0A1I8Q9R6 A0A3B0JTI7 A0A3B0JQS4 A0A3B0J3P6 A0A1I8Q9U8 A0A2J7PZI8 A0A0J9RWR5 A0A0J9RVW0 Q2M199 A0A0R3P2W1 A0A151XH84 A0A0K8TVP3 A0A0K8WGH1 U4UNG3 W8C0N9 A0A034V4C9 A0A067RBW8 A0A1W4WQT9 N6TLR6 B4QKK3 B4GUE9 A0A0A1WTW3 A0A0A1XD75 A0A2A3EFG1 E0VBA6 E2BF07 D2A085 A0A0J7L195 E9IDI3
Ontologies
GO
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.996172
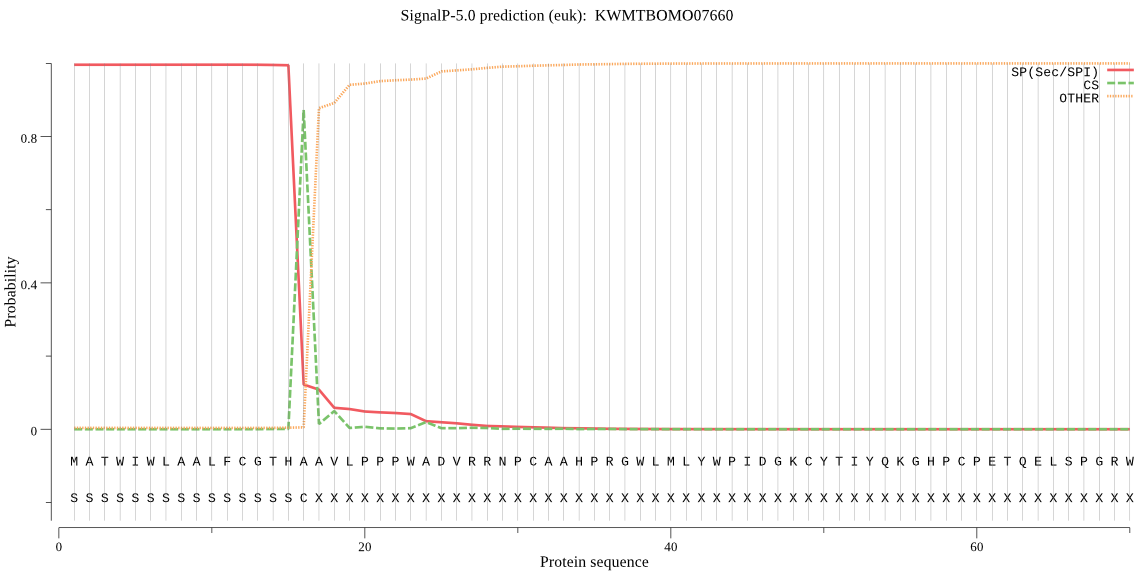
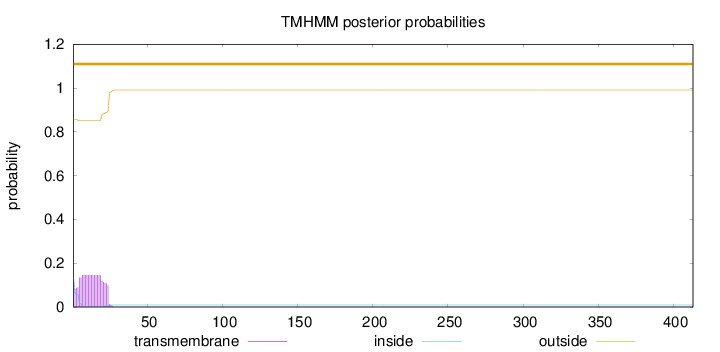
Length:
413
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.99811
Exp number, first 60 AAs:
2.99811
Total prob of N-in:
0.14332
outside
1 - 413
Population Genetic Test Statistics
Pi
253.061837
Theta
168.21583
Tajima's D
1.825442
CLR
0.025589
CSRT
0.855657217139143
Interpretation
Uncertain
