Gene
KWMTBOMO07659
Pre Gene Modal
BGIBMGA001290
Annotation
PREDICTED:_TRAF6_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.436
Sequence
CDS
ATGGACCGAAACAGAGCTAAAGATGCTTCAGAAGGGATCGCTCCTATGGGCATAACTGCATTAAATGAGACAGTTTTAAGTAACCAGCCAGAGTCACGATACGAATGTCCTGTTTGTTTAAACTGGCTTAGAGATCCAGTAATCACAACATGCGGACACAAGTTTTGTAAAAGCTGCATTACGTCATGGCTACAAAACAGTGGTCACTGTCCAATCGATAATATTAATCTAAGTATGAAAGTGGATATATTTCCTGATAACTACACTAAAAGAGAAATACAAGAACAAAGAATGAACTGTCCATTTTCAGCTAAAGGATGTACAGTAAAAGTAACCCCCTTAGACTTAGATGCACACATAGCTGTGTGCGAGTACAATCAACCGGAAACATCTTCACAACCAGAAATACACATCCCTTGCTCATTCAAGCCAGCCGGTTGTAAAGAAACATTTGATAGTCAAGATGAAATGAACCAACATTTGAATAATGATACCCAGAGTCATATGAATTTACTCATGAATGCATATTCAGAAATGAAAATCAATAATGATATGTCGAATGCTAATATGGACACTAAAGAACAAGAGGCGATGGCTCTGTGGGACGCACCTGATAAAGATGGCAATCAAACATCATCCCCACCACTTAACAATACCAGTGCACTAATACGAGCTCTATATGAACGTGTTGTCGTACTTGAACAAAGGAATCGCGAACAAGACATAGTCATAGCAAATATTTCGAAGCAACTCTCGGCTTTTGCCGTAGCCAAGATGAAACAAAACAATGAAATGTTACTGCGCTACTGCATGGGAAACTATGTGTGGAGAATCGATAACTTCAAAACGAGACTTGACGCCATGCTGAAGGACCATTACAAAATGTTGTACAGCCCTGGATTCTATACATCACCCAACGGTTATAGATTTTGTGTCCGGCTGAACATTTCACCGCAGAACACCCACTTCTTCGCTCTGCACGTGCACTTAATGAAGACCGAAAACGACGACTGCCTCGAATGGCCGTTCAACGGCAGGATATCGTTCGTCCTGGTCAACCAGGTCTACCCGGAGCTGTCCCAGCGCGACACGATGATGTCCAACACGAACTTGAAGGCGTTCAGTAAGCCCGCCACGGAGATATGCGTGCGAGGATTCGGCTACACGGAGTACGCTGTCCTCGGTGACGTCATTCGCAACGGCTTTGTCAAGGACGACGTTCTCATCATCAGAGTCAACATCAAATGCGTATGA
Protein
MDRNRAKDASEGIAPMGITALNETVLSNQPESRYECPVCLNWLRDPVITTCGHKFCKSCITSWLQNSGHCPIDNINLSMKVDIFPDNYTKREIQEQRMNCPFSAKGCTVKVTPLDLDAHIAVCEYNQPETSSQPEIHIPCSFKPAGCKETFDSQDEMNQHLNNDTQSHMNLLMNAYSEMKINNDMSNANMDTKEQEAMALWDAPDKDGNQTSSPPLNNTSALIRALYERVVVLEQRNREQDIVIANISKQLSAFAVAKMKQNNEMLLRYCMGNYVWRIDNFKTRLDAMLKDHYKMLYSPGFYTSPNGYRFCVRLNISPQNTHFFALHVHLMKTENDDCLEWPFNGRISFVLVNQVYPELSQRDTMMSNTNLKAFSKPATEICVRGFGYTEYAVLGDVIRNGFVKDDVLIIRVNIKCV
Summary
Uniprot
A0A2H1V3Z4
A0A2A4K7T0
A0A1E1W4E6
E9JEJ3
A0A212ES92
A0A3S2NS52
+ More
A0A194PDM7 A0A194QZX3 D2A1X0 A0A1W4W6B8 A0A1Y1NJY0 N6T883 A0A088AT24 A0A310S4W2 A0A2A3EID5 A0A1B6C0K9 A0A1B6CRS1 A0A0A1X2C8 A0A1B6J2U8 V5GZ69 A0A1B6FGM2 A0A1B6GMD4 A0A034W321 A0A0K8V072 W8C0H6 A0A154PPY8 A0A1L8DLN5 A0A1L8DMJ9 A0A0L0C3G6 A0A067RFW6 A0A0M4EM22 B4Q0E8 A0A2J7R2R9 A0A1I8M8V2 A0A182QDD0 A0A182WRK7 A0A182U9W8 A0A182RNS2 B3NX05 A0A182PKL6 Q9UAC4 A0A182ULZ7 A0A2M4AHW9 W5J6T9 A0A2M3Z6H8 A0A182FRD8 Q9W3I9 A0A2M4BRV7 B4JNP8 T1DRM4 A0A084VJT9 A0A1W4VXD1 A0A023EYV6 A0A0C9PK53 A0A182SJ79 A0A182JV48 B4L8G7 E2C776 A0A182J703 A0A182Y499 A0A182W055 A0A182LFV0 B4M202 A0A182MKP3 A0A1A9X565 A0A182N649 A0A0V0GBN6 A0A1B0G833 A0A3B0K888 Q29HD1 A0A1I8P8I3 Q9XYQ9 Q7PR29 A0A1A9VWT0 A0A182HU69 A0A1B0A171 A0A1A9XA24 A0A1B0AN13 A0A0L7REJ8 A0A195FJU3 A0A195E467 A0A158NXB6 B3MRL9 B4MSF1 A0A026WE70 A0A195BU78 F4WUP3 A0A195D350 T1HRR6 B4GXW7 A0A1B0DQ91 A0A151WXT0 A0A1B0GI36 A0A0A9WW92 A0A182TJJ4 B4IKY0 E0VE99 A0A2R7VZ10 A0A1B6MM67
A0A194PDM7 A0A194QZX3 D2A1X0 A0A1W4W6B8 A0A1Y1NJY0 N6T883 A0A088AT24 A0A310S4W2 A0A2A3EID5 A0A1B6C0K9 A0A1B6CRS1 A0A0A1X2C8 A0A1B6J2U8 V5GZ69 A0A1B6FGM2 A0A1B6GMD4 A0A034W321 A0A0K8V072 W8C0H6 A0A154PPY8 A0A1L8DLN5 A0A1L8DMJ9 A0A0L0C3G6 A0A067RFW6 A0A0M4EM22 B4Q0E8 A0A2J7R2R9 A0A1I8M8V2 A0A182QDD0 A0A182WRK7 A0A182U9W8 A0A182RNS2 B3NX05 A0A182PKL6 Q9UAC4 A0A182ULZ7 A0A2M4AHW9 W5J6T9 A0A2M3Z6H8 A0A182FRD8 Q9W3I9 A0A2M4BRV7 B4JNP8 T1DRM4 A0A084VJT9 A0A1W4VXD1 A0A023EYV6 A0A0C9PK53 A0A182SJ79 A0A182JV48 B4L8G7 E2C776 A0A182J703 A0A182Y499 A0A182W055 A0A182LFV0 B4M202 A0A182MKP3 A0A1A9X565 A0A182N649 A0A0V0GBN6 A0A1B0G833 A0A3B0K888 Q29HD1 A0A1I8P8I3 Q9XYQ9 Q7PR29 A0A1A9VWT0 A0A182HU69 A0A1B0A171 A0A1A9XA24 A0A1B0AN13 A0A0L7REJ8 A0A195FJU3 A0A195E467 A0A158NXB6 B3MRL9 B4MSF1 A0A026WE70 A0A195BU78 F4WUP3 A0A195D350 T1HRR6 B4GXW7 A0A1B0DQ91 A0A151WXT0 A0A1B0GI36 A0A0A9WW92 A0A182TJJ4 B4IKY0 E0VE99 A0A2R7VZ10 A0A1B6MM67
Pubmed
19121390
21040523
22118469
26354079
18362917
19820115
+ More
28004739 23537049 25830018 25348373 24495485 26108605 24845553 17994087 17550304 25315136 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24438588 25474469 20798317 25244985 20966253 15632085 10021364 12364791 21347285 24508170 30249741 21719571 25401762 26823975 20566863
28004739 23537049 25830018 25348373 24495485 26108605 24845553 17994087 17550304 25315136 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24438588 25474469 20798317 25244985 20966253 15632085 10021364 12364791 21347285 24508170 30249741 21719571 25401762 26823975 20566863
EMBL
ODYU01000571
SOQ35563.1
NWSH01000056
PCG80129.1
GDQN01009196
JAT81858.1
+ More
BABH01001343 BABH01001344 BABH01001345 GQ426310 ADM32536.1 AGBW02012839 OWR44362.1 RSAL01000275 RVE43117.1 KQ459606 KPI91118.1 KQ460947 KPJ10525.1 KQ971338 EFA02851.1 GEZM01001416 GEZM01001415 GEZM01001414 GEZM01001413 GEZM01001410 GEZM01001409 GEZM01001407 GEZM01001405 GEZM01001404 GEZM01001403 GEZM01001401 GEZM01001399 JAV97818.1 APGK01047844 KB741091 KB632368 ENN73928.1 ERL93680.1 KQ773364 OAD52384.1 KZ288254 PBC30761.1 GEDC01030247 GEDC01020642 GEDC01018876 GEDC01009232 JAS07051.1 JAS16656.1 JAS18422.1 JAS28066.1 GEDC01021180 JAS16118.1 GBXI01009030 JAD05262.1 GECU01014197 JAS93509.1 GALX01001264 JAB67202.1 GECZ01020485 JAS49284.1 GECZ01006171 JAS63598.1 GAKP01010432 JAC48520.1 GDHF01020078 JAI32236.1 GAMC01007419 JAB99136.1 KQ435028 KZC13963.1 GFDF01006706 JAV07378.1 GFDF01006398 JAV07686.1 JRES01000955 KNC26801.1 KK852491 KDR22761.1 CP012528 ALC48671.1 CM000162 EDX02285.1 NEVH01007828 PNF35131.1 AXCN02000196 CH954180 EDV46834.1 AF079838 AAD47895.1 GGFK01007001 MBW40322.1 ADMH02002131 ETN58489.1 GGFM01003381 MBW24132.1 AE014298 AY047501 AAF46338.1 AAK77233.1 GGFJ01006377 MBW55518.1 CH916371 EDV92341.1 GAMD01000585 JAB01006.1 ATLV01013870 KE524906 KFB38233.1 GBBI01004345 JAC14367.1 GBYB01001393 GBYB01001395 JAG71160.1 JAG71162.1 CH933815 EDW07942.2 GL453340 EFN76189.1 CH940651 EDW65706.2 AXCM01000845 GECL01001367 JAP04757.1 CCAG010000157 OUUW01000016 SPP88882.1 CH379064 EAL31827.1 AF119793 AAD34345.1 AAAB01008859 EAA07925.6 APCN01000610 JXJN01000694 KQ414609 KOC69259.1 KQ981523 KYN40274.1 KQ979657 KYN19980.1 ADTU01029620 CH902622 EDV34424.1 CH963851 EDW75040.2 KK107250 QOIP01000002 EZA54362.1 RLU25883.1 KQ976408 KYM91174.1 GL888375 EGI62040.1 KQ976885 KYN07335.1 ACPB03022414 CH479196 EDW27594.1 AJVK01018951 KQ982661 KYQ52663.1 AJWK01011243 AJWK01011244 AJWK01011245 GBHO01043398 GBHO01034516 GBHO01034515 GBHO01034514 GBHO01034513 GBHO01028922 GBHO01025204 GBHO01025181 GBHO01025177 GBHO01019954 GBHO01001773 GDHC01016793 GDHC01001727 GDHC01001649 GDHC01001200 JAG00206.1 JAG09088.1 JAG09089.1 JAG09090.1 JAG09091.1 JAG14682.1 JAG18400.1 JAG18423.1 JAG18427.1 JAG23650.1 JAG41831.1 JAQ01836.1 JAQ16902.1 JAQ16980.1 JAQ17429.1 CH480859 EDW52751.1 DS235088 EEB11705.1 KK854182 PTY12717.1 GEBQ01002943 JAT37034.1
BABH01001343 BABH01001344 BABH01001345 GQ426310 ADM32536.1 AGBW02012839 OWR44362.1 RSAL01000275 RVE43117.1 KQ459606 KPI91118.1 KQ460947 KPJ10525.1 KQ971338 EFA02851.1 GEZM01001416 GEZM01001415 GEZM01001414 GEZM01001413 GEZM01001410 GEZM01001409 GEZM01001407 GEZM01001405 GEZM01001404 GEZM01001403 GEZM01001401 GEZM01001399 JAV97818.1 APGK01047844 KB741091 KB632368 ENN73928.1 ERL93680.1 KQ773364 OAD52384.1 KZ288254 PBC30761.1 GEDC01030247 GEDC01020642 GEDC01018876 GEDC01009232 JAS07051.1 JAS16656.1 JAS18422.1 JAS28066.1 GEDC01021180 JAS16118.1 GBXI01009030 JAD05262.1 GECU01014197 JAS93509.1 GALX01001264 JAB67202.1 GECZ01020485 JAS49284.1 GECZ01006171 JAS63598.1 GAKP01010432 JAC48520.1 GDHF01020078 JAI32236.1 GAMC01007419 JAB99136.1 KQ435028 KZC13963.1 GFDF01006706 JAV07378.1 GFDF01006398 JAV07686.1 JRES01000955 KNC26801.1 KK852491 KDR22761.1 CP012528 ALC48671.1 CM000162 EDX02285.1 NEVH01007828 PNF35131.1 AXCN02000196 CH954180 EDV46834.1 AF079838 AAD47895.1 GGFK01007001 MBW40322.1 ADMH02002131 ETN58489.1 GGFM01003381 MBW24132.1 AE014298 AY047501 AAF46338.1 AAK77233.1 GGFJ01006377 MBW55518.1 CH916371 EDV92341.1 GAMD01000585 JAB01006.1 ATLV01013870 KE524906 KFB38233.1 GBBI01004345 JAC14367.1 GBYB01001393 GBYB01001395 JAG71160.1 JAG71162.1 CH933815 EDW07942.2 GL453340 EFN76189.1 CH940651 EDW65706.2 AXCM01000845 GECL01001367 JAP04757.1 CCAG010000157 OUUW01000016 SPP88882.1 CH379064 EAL31827.1 AF119793 AAD34345.1 AAAB01008859 EAA07925.6 APCN01000610 JXJN01000694 KQ414609 KOC69259.1 KQ981523 KYN40274.1 KQ979657 KYN19980.1 ADTU01029620 CH902622 EDV34424.1 CH963851 EDW75040.2 KK107250 QOIP01000002 EZA54362.1 RLU25883.1 KQ976408 KYM91174.1 GL888375 EGI62040.1 KQ976885 KYN07335.1 ACPB03022414 CH479196 EDW27594.1 AJVK01018951 KQ982661 KYQ52663.1 AJWK01011243 AJWK01011244 AJWK01011245 GBHO01043398 GBHO01034516 GBHO01034515 GBHO01034514 GBHO01034513 GBHO01028922 GBHO01025204 GBHO01025181 GBHO01025177 GBHO01019954 GBHO01001773 GDHC01016793 GDHC01001727 GDHC01001649 GDHC01001200 JAG00206.1 JAG09088.1 JAG09089.1 JAG09090.1 JAG09091.1 JAG14682.1 JAG18400.1 JAG18423.1 JAG18427.1 JAG23650.1 JAG41831.1 JAQ01836.1 JAQ16902.1 JAQ16980.1 JAQ17429.1 CH480859 EDW52751.1 DS235088 EEB11705.1 KK854182 PTY12717.1 GEBQ01002943 JAT37034.1
Proteomes
UP000218220
UP000005204
UP000007151
UP000283053
UP000053268
UP000053240
+ More
UP000007266 UP000192223 UP000019118 UP000030742 UP000005203 UP000242457 UP000076502 UP000037069 UP000027135 UP000092553 UP000002282 UP000235965 UP000095301 UP000075886 UP000076407 UP000075902 UP000075900 UP000008711 UP000075885 UP000075903 UP000000673 UP000069272 UP000000803 UP000001070 UP000030765 UP000192221 UP000075901 UP000075881 UP000009192 UP000008237 UP000075880 UP000076408 UP000075920 UP000075882 UP000008792 UP000075883 UP000091820 UP000075884 UP000092444 UP000268350 UP000001819 UP000095300 UP000007062 UP000078200 UP000075840 UP000092445 UP000092443 UP000092460 UP000053825 UP000078541 UP000078492 UP000005205 UP000007801 UP000007798 UP000053097 UP000279307 UP000078540 UP000007755 UP000078542 UP000015103 UP000008744 UP000092462 UP000075809 UP000092461 UP000001292 UP000009046
UP000007266 UP000192223 UP000019118 UP000030742 UP000005203 UP000242457 UP000076502 UP000037069 UP000027135 UP000092553 UP000002282 UP000235965 UP000095301 UP000075886 UP000076407 UP000075902 UP000075900 UP000008711 UP000075885 UP000075903 UP000000673 UP000069272 UP000000803 UP000001070 UP000030765 UP000192221 UP000075901 UP000075881 UP000009192 UP000008237 UP000075880 UP000076408 UP000075920 UP000075882 UP000008792 UP000075883 UP000091820 UP000075884 UP000092444 UP000268350 UP000001819 UP000095300 UP000007062 UP000078200 UP000075840 UP000092445 UP000092443 UP000092460 UP000053825 UP000078541 UP000078492 UP000005205 UP000007801 UP000007798 UP000053097 UP000279307 UP000078540 UP000007755 UP000078542 UP000015103 UP000008744 UP000092462 UP000075809 UP000092461 UP000001292 UP000009046
Interpro
SUPFAM
SSF49599
SSF49599
Gene 3D
ProteinModelPortal
A0A2H1V3Z4
A0A2A4K7T0
A0A1E1W4E6
E9JEJ3
A0A212ES92
A0A3S2NS52
+ More
A0A194PDM7 A0A194QZX3 D2A1X0 A0A1W4W6B8 A0A1Y1NJY0 N6T883 A0A088AT24 A0A310S4W2 A0A2A3EID5 A0A1B6C0K9 A0A1B6CRS1 A0A0A1X2C8 A0A1B6J2U8 V5GZ69 A0A1B6FGM2 A0A1B6GMD4 A0A034W321 A0A0K8V072 W8C0H6 A0A154PPY8 A0A1L8DLN5 A0A1L8DMJ9 A0A0L0C3G6 A0A067RFW6 A0A0M4EM22 B4Q0E8 A0A2J7R2R9 A0A1I8M8V2 A0A182QDD0 A0A182WRK7 A0A182U9W8 A0A182RNS2 B3NX05 A0A182PKL6 Q9UAC4 A0A182ULZ7 A0A2M4AHW9 W5J6T9 A0A2M3Z6H8 A0A182FRD8 Q9W3I9 A0A2M4BRV7 B4JNP8 T1DRM4 A0A084VJT9 A0A1W4VXD1 A0A023EYV6 A0A0C9PK53 A0A182SJ79 A0A182JV48 B4L8G7 E2C776 A0A182J703 A0A182Y499 A0A182W055 A0A182LFV0 B4M202 A0A182MKP3 A0A1A9X565 A0A182N649 A0A0V0GBN6 A0A1B0G833 A0A3B0K888 Q29HD1 A0A1I8P8I3 Q9XYQ9 Q7PR29 A0A1A9VWT0 A0A182HU69 A0A1B0A171 A0A1A9XA24 A0A1B0AN13 A0A0L7REJ8 A0A195FJU3 A0A195E467 A0A158NXB6 B3MRL9 B4MSF1 A0A026WE70 A0A195BU78 F4WUP3 A0A195D350 T1HRR6 B4GXW7 A0A1B0DQ91 A0A151WXT0 A0A1B0GI36 A0A0A9WW92 A0A182TJJ4 B4IKY0 E0VE99 A0A2R7VZ10 A0A1B6MM67
A0A194PDM7 A0A194QZX3 D2A1X0 A0A1W4W6B8 A0A1Y1NJY0 N6T883 A0A088AT24 A0A310S4W2 A0A2A3EID5 A0A1B6C0K9 A0A1B6CRS1 A0A0A1X2C8 A0A1B6J2U8 V5GZ69 A0A1B6FGM2 A0A1B6GMD4 A0A034W321 A0A0K8V072 W8C0H6 A0A154PPY8 A0A1L8DLN5 A0A1L8DMJ9 A0A0L0C3G6 A0A067RFW6 A0A0M4EM22 B4Q0E8 A0A2J7R2R9 A0A1I8M8V2 A0A182QDD0 A0A182WRK7 A0A182U9W8 A0A182RNS2 B3NX05 A0A182PKL6 Q9UAC4 A0A182ULZ7 A0A2M4AHW9 W5J6T9 A0A2M3Z6H8 A0A182FRD8 Q9W3I9 A0A2M4BRV7 B4JNP8 T1DRM4 A0A084VJT9 A0A1W4VXD1 A0A023EYV6 A0A0C9PK53 A0A182SJ79 A0A182JV48 B4L8G7 E2C776 A0A182J703 A0A182Y499 A0A182W055 A0A182LFV0 B4M202 A0A182MKP3 A0A1A9X565 A0A182N649 A0A0V0GBN6 A0A1B0G833 A0A3B0K888 Q29HD1 A0A1I8P8I3 Q9XYQ9 Q7PR29 A0A1A9VWT0 A0A182HU69 A0A1B0A171 A0A1A9XA24 A0A1B0AN13 A0A0L7REJ8 A0A195FJU3 A0A195E467 A0A158NXB6 B3MRL9 B4MSF1 A0A026WE70 A0A195BU78 F4WUP3 A0A195D350 T1HRR6 B4GXW7 A0A1B0DQ91 A0A151WXT0 A0A1B0GI36 A0A0A9WW92 A0A182TJJ4 B4IKY0 E0VE99 A0A2R7VZ10 A0A1B6MM67
PDB
6MYD
E-value=1.70792e-21,
Score=254
Ontologies
PATHWAY
04013
MAPK signaling pathway - fly - Bombyx mori (domestic silkworm)
04120 Ubiquitin mediated proteolysis - Bombyx mori (domestic silkworm)
04140 Autophagy - animal - Bombyx mori (domestic silkworm)
04144 Endocytosis - Bombyx mori (domestic silkworm)
04214 Apoptosis - fly - Bombyx mori (domestic silkworm)
04120 Ubiquitin mediated proteolysis - Bombyx mori (domestic silkworm)
04140 Autophagy - animal - Bombyx mori (domestic silkworm)
04144 Endocytosis - Bombyx mori (domestic silkworm)
04214 Apoptosis - fly - Bombyx mori (domestic silkworm)
GO
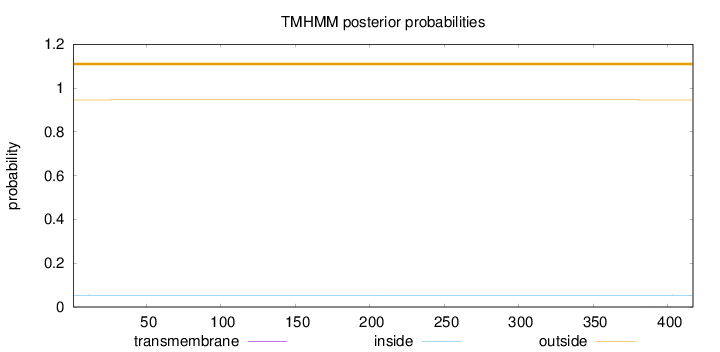
Topology
Length:
417
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01168
Exp number, first 60 AAs:
0.00122
Total prob of N-in:
0.05405
outside
1 - 417
Population Genetic Test Statistics
Pi
150.969224
Theta
187.860415
Tajima's D
-1.319139
CLR
0
CSRT
0.0887955602219889
Interpretation
Uncertain
