Gene
KWMTBOMO07657
Pre Gene Modal
BGIBMGA001291
Annotation
PREDICTED:_iduronate_2-sulfatase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.562 Nuclear Reliability : 1.008
Sequence
CDS
ATGCAGAGGCTCACAATGATTTATGTCGTAAATATAATATTGTTGAATGGTGATCGTGTACTTACAAGTGACGTAGAAACACCGAAAAATATCCTGTTTATTTTGATTGACGATTTGCGCCATCTCTCGGACAAAAAAGTTTATTTGCCTAACATCAACTTCTTAGGAAAAACTGGTGCTACCTTTAATAATGCGTTCGCTCAACAAGCCTTATGTGCGCCCAGTCGTAATTCCCTTCTAACAGGTCGTAGGCCCGACTCGCTTCGCTTGTACGACTTCTACAGCTACTGGCGCGATCGCAGTAACGGACAAGGGAATTTCACAACCATACCACAGTTCTTCAAAGAGCATGGATACGATACTTACTCCGTGGGAAAAGTGTTCCATCCTGGAAAAAGTTCGAACTTCACAGACGATTACCCGTACAGCTGGTCGGAGTATCCGTATCATCCCCCGACAGAAATGTACAAAGACGCTAAAGTGTGTCGAAATAAAAAAACGAAAAAACTAGAGAGAAATCTAATTTGTCCAGTTTCTGTTAAAAGACAACCAGGACAATCTCTACCAGACTTGCAGTCGTTGGATTACGCTATTGATTTCCTGAAAAAGAGAAACGGGAGTAAGCCGTTTTTCTTAGCAATCGGATTTCACAAGCCGCATATACCATTGAAGTTTCCAAAGGAATATTTAAAACAAATGCCAATTAGCAAAGTGCATAGACCTAAAGAACCGAACATTCCCAAGGACATGCCTTTGGTGTCCTGGCACCCGTGGACGGACGTACGGAAACGCGATGACATTCGCCGCTTAAACATCACATTTCCTTTCGGCGTGATGCCCACTAAATGGACGTTGAAGATACGGCAGAGTTACTACGCGGCGGCTTTGTACATTGACGAATTGATTGGGATCCTGTTAAGCTATGTGGACATGCAGAAAACTATAATTGTACTGACAAGTGATCATGGTTGGTCACTCGGCGAAAACGGACTTTGGGCAAAATACAGTAACTTCGATTACGCTCTTAAAGTTCCATTAATATTCAAATCTCCCAAACTGATTCCAACTGTTGTCCATGAACCAGTCGAATTAATAGACATTTTCCCGACCCTGGTTGATCTTACCAAATTATCTGACGAAATACCCAAATGTCTGAACCACAAAGACACGTCACAGCTTTGTTTCGAAGGAAAAAGTTTAGTTCCTTTCATTGAAAATAATAGTAATGGACTTGAAGCTTTTGCAATAAGTCAGTGCCCGCGACCGAGTGTGTATCCGCAGAAAAATTCGGACAAACCACGTTTGAAGGACATCACTATTATGGGTTACAGCATCAGAACGAAAAGATACAGATACACAGAATGGATATCGTTTAATAACACGCTCTTTACAAAAAATTGGAACAATAATTATGGCATAGAACTGTACGATCACATTATTGATCCAATCGAATCTAAAAACTTATTTTTGGTCTCAAAATACAAAAATATTGCAAAGGTCTTATCTATACGATTAAGATCTAGTGTTTATGTCATTGTTTAA
Protein
MQRLTMIYVVNIILLNGDRVLTSDVETPKNILFILIDDLRHLSDKKVYLPNINFLGKTGATFNNAFAQQALCAPSRNSLLTGRRPDSLRLYDFYSYWRDRSNGQGNFTTIPQFFKEHGYDTYSVGKVFHPGKSSNFTDDYPYSWSEYPYHPPTEMYKDAKVCRNKKTKKLERNLICPVSVKRQPGQSLPDLQSLDYAIDFLKKRNGSKPFFLAIGFHKPHIPLKFPKEYLKQMPISKVHRPKEPNIPKDMPLVSWHPWTDVRKRDDIRRLNITFPFGVMPTKWTLKIRQSYYAAALYIDELIGILLSYVDMQKTIIVLTSDHGWSLGENGLWAKYSNFDYALKVPLIFKSPKLIPTVVHEPVELIDIFPTLVDLTKLSDEIPKCLNHKDTSQLCFEGKSLVPFIENNSNGLEAFAISQCPRPSVYPQKNSDKPRLKDITIMGYSIRTKRYRYTEWISFNNTLFTKNWNNNYGIELYDHIIDPIESKNLFLVSKYKNIAKVLSIRLRSSVYVIV
Summary
Uniprot
H9IVL1
A0A2H1VA53
A0A194PCT6
A0A212ESD8
A0A0L7LHC0
A0A2A4K7B3
+ More
A0A194QYA5 A0A1B0C9E5 D6WDU1 A0A182QHX5 A0A182NJQ6 A0A1B0DI78 A0A1Y1KM57 A0A1B6MUH9 A0A182Y253 A0A182WKZ7 A0A2J7Q121 B0WCM5 A0A067RAA8 Q1WJM2 E0W0K2 A0A146LCA1 A0A182L376 A0A182LRM1 A0A182XMK9 Q7Q6L4 A0A1Q3FMZ0 A0A1Q3FMT6 A0A182VAD6 Q1WJL8 A0A182HWG8 A0A182JEH9 A0A1W7R8L7 A0A084WJP8 A0A1I8MGT1 A0A182RN00 A0A182JRP6 N6U729 A0A0L0BMP7 W5J2U8 A0A336M9N0 A0A1Q3FMK0 A0A1S4EZP7 A0A182FHV0 A0A1A9VHE0 A0A1I8QFC1 A8WGX6 B4KZK8 B4LF09 B3M833 A0A1L8F7I5 H3AV81 A0A1B0FKZ6 A0A0A9XN97 A0A1A9Z969 Q29DQ4 B4H781 A0A3P9I2L1 B4K2U7 B4J3H2 A0A3Q3JMH7 A0A1A9XSE5 A0A3P9I2T9 A0A1A9WG07 B4PGQ4 A0A3P9LF10 A0A3B0JYU9 A0A3Q1J2T4 A0A3Q3JEU1 A0A3P9MM54 A0A1B0BIL6 A0A091RKS9 A0A3Q3JFE8 V9KJ45 A0A3B3CCB1 A0A3Q3JML2 V9KIC9 H2LZB9 A0A034WLL5 A0A2J7Q126 A0A091KK97 A0A3P8QQG2 A0A3P8QS74 A0A1W4VP74 H0Z2M6 A0A3P9BBD6 A0A3Q2YP10 K7J123 A0A091VRR3 B5RJ62 Q4V5Y6 Q9VZP8 Q4V5R8 B4HTR6 B4QPI8 A0A3B4ENN1 A0A091J449 A0A0K8V4K8 A0A0M3QVF8 A0A3S3PJX6
A0A194QYA5 A0A1B0C9E5 D6WDU1 A0A182QHX5 A0A182NJQ6 A0A1B0DI78 A0A1Y1KM57 A0A1B6MUH9 A0A182Y253 A0A182WKZ7 A0A2J7Q121 B0WCM5 A0A067RAA8 Q1WJM2 E0W0K2 A0A146LCA1 A0A182L376 A0A182LRM1 A0A182XMK9 Q7Q6L4 A0A1Q3FMZ0 A0A1Q3FMT6 A0A182VAD6 Q1WJL8 A0A182HWG8 A0A182JEH9 A0A1W7R8L7 A0A084WJP8 A0A1I8MGT1 A0A182RN00 A0A182JRP6 N6U729 A0A0L0BMP7 W5J2U8 A0A336M9N0 A0A1Q3FMK0 A0A1S4EZP7 A0A182FHV0 A0A1A9VHE0 A0A1I8QFC1 A8WGX6 B4KZK8 B4LF09 B3M833 A0A1L8F7I5 H3AV81 A0A1B0FKZ6 A0A0A9XN97 A0A1A9Z969 Q29DQ4 B4H781 A0A3P9I2L1 B4K2U7 B4J3H2 A0A3Q3JMH7 A0A1A9XSE5 A0A3P9I2T9 A0A1A9WG07 B4PGQ4 A0A3P9LF10 A0A3B0JYU9 A0A3Q1J2T4 A0A3Q3JEU1 A0A3P9MM54 A0A1B0BIL6 A0A091RKS9 A0A3Q3JFE8 V9KJ45 A0A3B3CCB1 A0A3Q3JML2 V9KIC9 H2LZB9 A0A034WLL5 A0A2J7Q126 A0A091KK97 A0A3P8QQG2 A0A3P8QS74 A0A1W4VP74 H0Z2M6 A0A3P9BBD6 A0A3Q2YP10 K7J123 A0A091VRR3 B5RJ62 Q4V5Y6 Q9VZP8 Q4V5R8 B4HTR6 B4QPI8 A0A3B4ENN1 A0A091J449 A0A0K8V4K8 A0A0M3QVF8 A0A3S3PJX6
Pubmed
19121390
26354079
22118469
26227816
18362917
19820115
+ More
28004739 25244985 24845553 16606844 20566863 26823975 20966253 12364791 14747013 17210077 24438588 25315136 23537049 26108605 20920257 23761445 17510324 20431018 17994087 27762356 9215903 25401762 15632085 17554307 17550304 24402279 29451363 25348373 20360741 25186727 20075255 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249
28004739 25244985 24845553 16606844 20566863 26823975 20966253 12364791 14747013 17210077 24438588 25315136 23537049 26108605 20920257 23761445 17510324 20431018 17994087 27762356 9215903 25401762 15632085 17554307 17550304 24402279 29451363 25348373 20360741 25186727 20075255 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249
EMBL
BABH01001346
ODYU01001210
SOQ37114.1
KQ459606
KPI91116.1
AGBW02012839
+ More
OWR44364.1 JTDY01001137 KOB74764.1 NWSH01000056 PCG80127.1 KQ460947 KPJ10523.1 AJWK01002262 KQ971323 EFA00827.1 AXCN02000515 AJVK01062130 GEZM01081385 GEZM01081383 JAV61668.1 GEBQ01000409 JAT39568.1 NEVH01019960 PNF22275.1 DS231889 EDS43676.1 KK852782 KDR16620.1 DQ230893 ABD94312.1 DS235860 EEB19158.1 GDHC01013340 JAQ05289.1 AXCM01004096 AAAB01008960 EAA11898.4 GFDL01006147 JAV28898.1 GFDL01006146 JAV28899.1 DQ230896 ABD94316.1 APCN01001491 GEHC01000175 JAV47470.1 ATLV01024046 KE525348 KFB50442.1 APGK01046894 APGK01046895 KB741069 KB632402 ENN74397.1 ERL94938.1 JRES01001646 KNC21208.1 ADMH02002165 ETN58116.1 UFQT01000765 SSX27052.1 GFDL01006293 JAV28752.1 AAMC01035134 BC154891 AAI54892.1 CH933809 EDW17935.2 CH940647 EDW69178.2 CH902618 EDV39941.1 CM004480 OCT67552.1 AFYH01036636 AFYH01036637 CCAG010008634 GBHO01021417 JAG22187.1 CH379070 EAL30360.2 CH479217 EDW33687.1 CH920042 EDV90423.1 CH916366 EDV96174.1 CM000159 EDW93272.1 OUUW01000012 SPP87224.1 JXJN01014988 KK801572 KFQ29142.1 JW865475 AFO97992.1 JW865181 AFO97698.1 GAKP01003735 JAC55217.1 PNF22276.1 KK743840 KFP39600.1 ABQF01018676 ABQF01018677 ABQF01018678 ABQF01018679 ABQF01018680 ABQF01018681 ABQF01018682 ABQF01018683 KL411262 KFR05198.1 BT044336 ACH92401.1 BT022520 AAY54936.1 AE014296 AAF47771.1 BT022588 AAY55004.1 CH480817 EDW50337.1 CM000363 CM002912 EDX09081.1 KMY97361.1 KL218558 KFP06586.1 GDHF01028953 GDHF01027423 GDHF01018452 JAI23361.1 JAI24891.1 JAI33862.1 CP012524 ALC42303.1 NCKU01001010 NCKU01000827 NCKU01000817 RWS13367.1 RWS13996.1 RWS14036.1
OWR44364.1 JTDY01001137 KOB74764.1 NWSH01000056 PCG80127.1 KQ460947 KPJ10523.1 AJWK01002262 KQ971323 EFA00827.1 AXCN02000515 AJVK01062130 GEZM01081385 GEZM01081383 JAV61668.1 GEBQ01000409 JAT39568.1 NEVH01019960 PNF22275.1 DS231889 EDS43676.1 KK852782 KDR16620.1 DQ230893 ABD94312.1 DS235860 EEB19158.1 GDHC01013340 JAQ05289.1 AXCM01004096 AAAB01008960 EAA11898.4 GFDL01006147 JAV28898.1 GFDL01006146 JAV28899.1 DQ230896 ABD94316.1 APCN01001491 GEHC01000175 JAV47470.1 ATLV01024046 KE525348 KFB50442.1 APGK01046894 APGK01046895 KB741069 KB632402 ENN74397.1 ERL94938.1 JRES01001646 KNC21208.1 ADMH02002165 ETN58116.1 UFQT01000765 SSX27052.1 GFDL01006293 JAV28752.1 AAMC01035134 BC154891 AAI54892.1 CH933809 EDW17935.2 CH940647 EDW69178.2 CH902618 EDV39941.1 CM004480 OCT67552.1 AFYH01036636 AFYH01036637 CCAG010008634 GBHO01021417 JAG22187.1 CH379070 EAL30360.2 CH479217 EDW33687.1 CH920042 EDV90423.1 CH916366 EDV96174.1 CM000159 EDW93272.1 OUUW01000012 SPP87224.1 JXJN01014988 KK801572 KFQ29142.1 JW865475 AFO97992.1 JW865181 AFO97698.1 GAKP01003735 JAC55217.1 PNF22276.1 KK743840 KFP39600.1 ABQF01018676 ABQF01018677 ABQF01018678 ABQF01018679 ABQF01018680 ABQF01018681 ABQF01018682 ABQF01018683 KL411262 KFR05198.1 BT044336 ACH92401.1 BT022520 AAY54936.1 AE014296 AAF47771.1 BT022588 AAY55004.1 CH480817 EDW50337.1 CM000363 CM002912 EDX09081.1 KMY97361.1 KL218558 KFP06586.1 GDHF01028953 GDHF01027423 GDHF01018452 JAI23361.1 JAI24891.1 JAI33862.1 CP012524 ALC42303.1 NCKU01001010 NCKU01000827 NCKU01000817 RWS13367.1 RWS13996.1 RWS14036.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000037510
UP000218220
UP000053240
+ More
UP000092461 UP000007266 UP000075886 UP000075884 UP000092462 UP000076408 UP000075920 UP000235965 UP000002320 UP000027135 UP000009046 UP000075882 UP000075883 UP000076407 UP000007062 UP000075903 UP000075840 UP000075880 UP000030765 UP000095301 UP000075900 UP000075881 UP000019118 UP000030742 UP000037069 UP000000673 UP000069272 UP000078200 UP000095300 UP000008143 UP000009192 UP000008792 UP000007801 UP000186698 UP000008672 UP000092444 UP000092445 UP000001819 UP000008744 UP000265200 UP000001070 UP000261600 UP000092443 UP000091820 UP000002282 UP000265180 UP000268350 UP000265040 UP000092460 UP000261560 UP000001038 UP000265100 UP000192221 UP000007754 UP000265160 UP000264820 UP000002358 UP000053283 UP000000803 UP000001292 UP000000304 UP000261460 UP000054308 UP000092553 UP000285301
UP000092461 UP000007266 UP000075886 UP000075884 UP000092462 UP000076408 UP000075920 UP000235965 UP000002320 UP000027135 UP000009046 UP000075882 UP000075883 UP000076407 UP000007062 UP000075903 UP000075840 UP000075880 UP000030765 UP000095301 UP000075900 UP000075881 UP000019118 UP000030742 UP000037069 UP000000673 UP000069272 UP000078200 UP000095300 UP000008143 UP000009192 UP000008792 UP000007801 UP000186698 UP000008672 UP000092444 UP000092445 UP000001819 UP000008744 UP000265200 UP000001070 UP000261600 UP000092443 UP000091820 UP000002282 UP000265180 UP000268350 UP000265040 UP000092460 UP000261560 UP000001038 UP000265100 UP000192221 UP000007754 UP000265160 UP000264820 UP000002358 UP000053283 UP000000803 UP000001292 UP000000304 UP000261460 UP000054308 UP000092553 UP000285301
Interpro
Gene 3D
ProteinModelPortal
H9IVL1
A0A2H1VA53
A0A194PCT6
A0A212ESD8
A0A0L7LHC0
A0A2A4K7B3
+ More
A0A194QYA5 A0A1B0C9E5 D6WDU1 A0A182QHX5 A0A182NJQ6 A0A1B0DI78 A0A1Y1KM57 A0A1B6MUH9 A0A182Y253 A0A182WKZ7 A0A2J7Q121 B0WCM5 A0A067RAA8 Q1WJM2 E0W0K2 A0A146LCA1 A0A182L376 A0A182LRM1 A0A182XMK9 Q7Q6L4 A0A1Q3FMZ0 A0A1Q3FMT6 A0A182VAD6 Q1WJL8 A0A182HWG8 A0A182JEH9 A0A1W7R8L7 A0A084WJP8 A0A1I8MGT1 A0A182RN00 A0A182JRP6 N6U729 A0A0L0BMP7 W5J2U8 A0A336M9N0 A0A1Q3FMK0 A0A1S4EZP7 A0A182FHV0 A0A1A9VHE0 A0A1I8QFC1 A8WGX6 B4KZK8 B4LF09 B3M833 A0A1L8F7I5 H3AV81 A0A1B0FKZ6 A0A0A9XN97 A0A1A9Z969 Q29DQ4 B4H781 A0A3P9I2L1 B4K2U7 B4J3H2 A0A3Q3JMH7 A0A1A9XSE5 A0A3P9I2T9 A0A1A9WG07 B4PGQ4 A0A3P9LF10 A0A3B0JYU9 A0A3Q1J2T4 A0A3Q3JEU1 A0A3P9MM54 A0A1B0BIL6 A0A091RKS9 A0A3Q3JFE8 V9KJ45 A0A3B3CCB1 A0A3Q3JML2 V9KIC9 H2LZB9 A0A034WLL5 A0A2J7Q126 A0A091KK97 A0A3P8QQG2 A0A3P8QS74 A0A1W4VP74 H0Z2M6 A0A3P9BBD6 A0A3Q2YP10 K7J123 A0A091VRR3 B5RJ62 Q4V5Y6 Q9VZP8 Q4V5R8 B4HTR6 B4QPI8 A0A3B4ENN1 A0A091J449 A0A0K8V4K8 A0A0M3QVF8 A0A3S3PJX6
A0A194QYA5 A0A1B0C9E5 D6WDU1 A0A182QHX5 A0A182NJQ6 A0A1B0DI78 A0A1Y1KM57 A0A1B6MUH9 A0A182Y253 A0A182WKZ7 A0A2J7Q121 B0WCM5 A0A067RAA8 Q1WJM2 E0W0K2 A0A146LCA1 A0A182L376 A0A182LRM1 A0A182XMK9 Q7Q6L4 A0A1Q3FMZ0 A0A1Q3FMT6 A0A182VAD6 Q1WJL8 A0A182HWG8 A0A182JEH9 A0A1W7R8L7 A0A084WJP8 A0A1I8MGT1 A0A182RN00 A0A182JRP6 N6U729 A0A0L0BMP7 W5J2U8 A0A336M9N0 A0A1Q3FMK0 A0A1S4EZP7 A0A182FHV0 A0A1A9VHE0 A0A1I8QFC1 A8WGX6 B4KZK8 B4LF09 B3M833 A0A1L8F7I5 H3AV81 A0A1B0FKZ6 A0A0A9XN97 A0A1A9Z969 Q29DQ4 B4H781 A0A3P9I2L1 B4K2U7 B4J3H2 A0A3Q3JMH7 A0A1A9XSE5 A0A3P9I2T9 A0A1A9WG07 B4PGQ4 A0A3P9LF10 A0A3B0JYU9 A0A3Q1J2T4 A0A3Q3JEU1 A0A3P9MM54 A0A1B0BIL6 A0A091RKS9 A0A3Q3JFE8 V9KJ45 A0A3B3CCB1 A0A3Q3JML2 V9KIC9 H2LZB9 A0A034WLL5 A0A2J7Q126 A0A091KK97 A0A3P8QQG2 A0A3P8QS74 A0A1W4VP74 H0Z2M6 A0A3P9BBD6 A0A3Q2YP10 K7J123 A0A091VRR3 B5RJ62 Q4V5Y6 Q9VZP8 Q4V5R8 B4HTR6 B4QPI8 A0A3B4ENN1 A0A091J449 A0A0K8V4K8 A0A0M3QVF8 A0A3S3PJX6
PDB
5FQL
E-value=1.48308e-116,
Score=1074
Ontologies
PATHWAY
GO
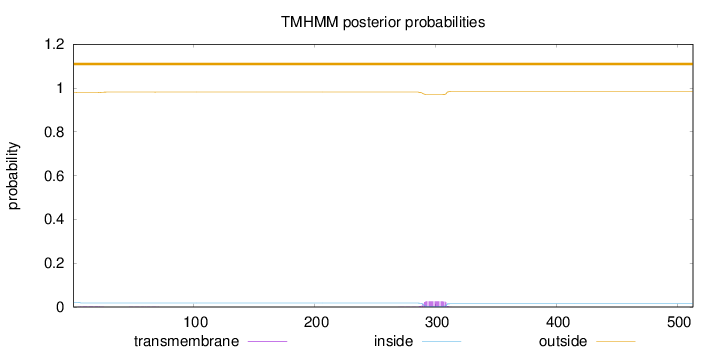
Topology
SignalP
Position: 1 - 22,
Likelihood: 0.656121
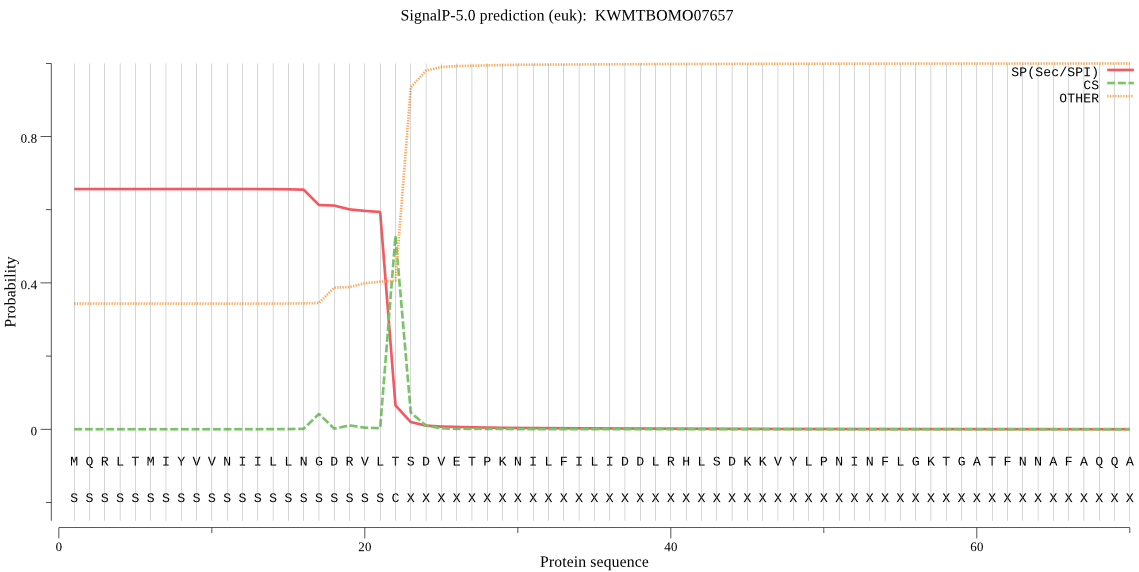
Length:
513
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.56829
Exp number, first 60 AAs:
0.04683
Total prob of N-in:
0.02070
outside
1 - 513
Population Genetic Test Statistics
Pi
157.575942
Theta
151.356831
Tajima's D
0.131834
CLR
0.146382
CSRT
0.402429878506075
Interpretation
Uncertain
