Gene
KWMTBOMO07656
Pre Gene Modal
BGIBMGA000854
Annotation
PREDICTED:_GDP-fucose_transporter_1_[Papilio_xuthus]
Full name
GDP-fucose transporter 1
Alternative Name
Neuronally altered carbohydrate
Solute carrier family 35 member C1 homolog
Solute carrier family 35 member C1 homolog
Location in the cell
PlasmaMembrane Reliability : 4.888
Sequence
CDS
ATGAAGTCTCAGCAATCGGAATTATGTTCTCTCTATACCAAGATATTCGTAGTGGTTTCTTGTTACTGGGTGGTGTCAATAGTGACAGTATTTGTTAATAAGACACTATTCAGCAGTCAAACAGTGGCTCTTGAGGCGCCTCTATTCATAACTTGGTTCCAGTGTATAATTTCATTCTCAATTTGTTTTACATTGAGTCAAAATGGTGGAGTTCCTGGTATATTTTCCTTTCCAAAAGGAACACCTTGGAGTATGGACGTCATGAAAAAGGTGATTCCACTGGCAATAATGTATACTTTGATGATAGCCACAAATAATTTGTGTTTGAAGCATGTAGGAGTACCGTTCTATTATGTAGGAAGATCTCTGACCACAGTCTTCAATGTTGTATTTAGCTGGATATTATTAAGACAGGCAACATCGTCCAGATGTATTTTGTGCTGCGCCTTCATTATATTCGGATTCTATCTTGGAGTTGATCAAGAAAACTTATTAGGGTCATTTTCCCTTATAGGAACAATCTATGGAGTGGTTGGTTCCCTCATGCTATCCTTGTACTCTATATTCACAAAGAAGGTTTTACCATATGTTAACCAAGAAGTATTGTTATTGTCGTACTACAATAATGCATACTCCATAATACTTTTCATACCGCTAATGTTAATCAACGGAGAAATGACAGTTTTGATGAACTACACTAATTTCCAAAGCACCTACTTCTGGTTTCAAATGGTAATAGGTGGTCTATGCGGTTTTGCAATAGGATACTGTACTTCTCTACAGATAAAGGTAACATCACCTCTGACACACAATATATCTGGCACAGCAAAGGCTTGTGCGCAAACGGTGATCGCGACGCAGTGGTACAACGAAAGTAAAAATGCGCTTTGGTGGACGTCCAATTTAATAGTACTAGCCAGTTCAGCTTTATACGCAAGATTCAAGCAGTTGGAGATGGAAGTTAAATCAAAGACGCCAGTGCCTGAAGAAAAACAGAGCTTAGTATAA
Protein
MKSQQSELCSLYTKIFVVVSCYWVVSIVTVFVNKTLFSSQTVALEAPLFITWFQCIISFSICFTLSQNGGVPGIFSFPKGTPWSMDVMKKVIPLAIMYTLMIATNNLCLKHVGVPFYYVGRSLTTVFNVVFSWILLRQATSSRCILCCAFIIFGFYLGVDQENLLGSFSLIGTIYGVVGSLMLSLYSIFTKKVLPYVNQEVLLLSYYNNAYSIILFIPLMLINGEMTVLMNYTNFQSTYFWFQMVIGGLCGFAIGYCTSLQIKVTSPLTHNISGTAKACAQTVIATQWYNESKNALWWTSNLIVLASSALYARFKQLEMEVKSKTPVPEEKQSLV
Summary
Description
Involved in GDP-fucose import from the cytoplasm into the Golgi lumen. Plays a major role in the fucosylation of N-glycans. Functions redundantly with Efr in the O-fucosylation of Notch, positively regulating Notch signaling.
Similarity
Belongs to the TPT transporter family. SLC35C subfamily.
Keywords
Complete proteome
Golgi apparatus
Membrane
Reference proteome
Sugar transport
Transmembrane
Transmembrane helix
Transport
Feature
chain GDP-fucose transporter 1
Uniprot
A0A2A4K7Z0
A0A212ESA0
A0A2H1V8F2
A0A194PJ54
A0A194QYI2
A0A1E1WLY0
+ More
H9IUC4 A0A336L574 K7J4T3 A0A336MX09 A0A0K8UBE2 S4PNA2 A0A232F149 A0A1B0CBG2 A0A182MRR5 A0A0C9RP07 A0A1L8DED5 A0A1L8DEZ0 A0A182VX57 A0A1I8MES2 A0A154PKX3 A0A088ASJ3 W8C917 A0A0L0CRU3 A0A1I8PRB0 A0A1Q3FB01 A0A0M9A9E3 A0A1S4FGL2 B0WS63 Q171Y2 A0A0M3QXP1 B4NAF1 B4M5Q2 A0A0K8TRY9 A0A2A3EDK5 U5EJR6 A0A067QUH2 A0A182G1I3 A0A3B0JQL9 B3M0D2 A0A1W4VHG1 Q298H3 A0A182QHB8 A0A182GCP6 B4K7W1 B4PT36 Q9VHT4 B3NZU7 B4HLM5 A0A084W1P0 A0A1A9WY54 A0A182NA86 A0A182VD60 A0A182R3N3 A0A182PA61 A0A182F8D0 A0A1A9UZ23 A0A182IVJ0 A0A182TU73 A0A182XI94 E2BEE2 A0A182LFB2 Q7QC10 A0A182HT81 B4JH27 D3TRG4 A0A1J1HXS6 A0A1A9Z9M3 A0A182JX79 A0A1B6E6Z6 A0A1B6K0M0 A0A2M4BTH8 A0A2M4BTP3 A0A2M4BTN7 W5JKP2 A0A1B6F945 A0A1A9XG71 A0A195C7J3 A0A1B0BQ30 A0A1B0GAV4 A0A158NMV8 A0A026W5L1 E2B0K8 A0A1B6LE31 N6SRR0 A0A195EWR2 J3JV05 A0A1S3DHQ6 A0A1Y1N0R0 A0A182SDQ1 F4WT95 A0A195BB45 A0A151WHH2 A0A151IYP0 A0A1W4XDX3 J9JP93 A0A182YKI2 D6X1P8 A0A2S2Q391 A0A0A9Z075
H9IUC4 A0A336L574 K7J4T3 A0A336MX09 A0A0K8UBE2 S4PNA2 A0A232F149 A0A1B0CBG2 A0A182MRR5 A0A0C9RP07 A0A1L8DED5 A0A1L8DEZ0 A0A182VX57 A0A1I8MES2 A0A154PKX3 A0A088ASJ3 W8C917 A0A0L0CRU3 A0A1I8PRB0 A0A1Q3FB01 A0A0M9A9E3 A0A1S4FGL2 B0WS63 Q171Y2 A0A0M3QXP1 B4NAF1 B4M5Q2 A0A0K8TRY9 A0A2A3EDK5 U5EJR6 A0A067QUH2 A0A182G1I3 A0A3B0JQL9 B3M0D2 A0A1W4VHG1 Q298H3 A0A182QHB8 A0A182GCP6 B4K7W1 B4PT36 Q9VHT4 B3NZU7 B4HLM5 A0A084W1P0 A0A1A9WY54 A0A182NA86 A0A182VD60 A0A182R3N3 A0A182PA61 A0A182F8D0 A0A1A9UZ23 A0A182IVJ0 A0A182TU73 A0A182XI94 E2BEE2 A0A182LFB2 Q7QC10 A0A182HT81 B4JH27 D3TRG4 A0A1J1HXS6 A0A1A9Z9M3 A0A182JX79 A0A1B6E6Z6 A0A1B6K0M0 A0A2M4BTH8 A0A2M4BTP3 A0A2M4BTN7 W5JKP2 A0A1B6F945 A0A1A9XG71 A0A195C7J3 A0A1B0BQ30 A0A1B0GAV4 A0A158NMV8 A0A026W5L1 E2B0K8 A0A1B6LE31 N6SRR0 A0A195EWR2 J3JV05 A0A1S3DHQ6 A0A1Y1N0R0 A0A182SDQ1 F4WT95 A0A195BB45 A0A151WHH2 A0A151IYP0 A0A1W4XDX3 J9JP93 A0A182YKI2 D6X1P8 A0A2S2Q391 A0A0A9Z075
Pubmed
22118469
26354079
19121390
20075255
23622113
28648823
+ More
25315136 24495485 26108605 17510324 17994087 26369729 24845553 26483478 15632085 17550304 10731132 12537572 19948734 22745127 24438588 20798317 20966253 12364791 14747013 17210077 20353571 20920257 23761445 21347285 24508170 30249741 23537049 22516182 28004739 21719571 25244985 18362917 19820115 25401762 26823975
25315136 24495485 26108605 17510324 17994087 26369729 24845553 26483478 15632085 17550304 10731132 12537572 19948734 22745127 24438588 20798317 20966253 12364791 14747013 17210077 20353571 20920257 23761445 21347285 24508170 30249741 23537049 22516182 28004739 21719571 25244985 18362917 19820115 25401762 26823975
EMBL
NWSH01000056
PCG80126.1
AGBW02012839
OWR44365.1
ODYU01001210
SOQ37115.1
+ More
KQ459606 KPI91115.1 KQ460947 KPJ10522.1 GDQN01003021 JAT88033.1 BABH01001346 UFQS01001768 UFQT01001768 SSX12281.1 SSX31732.1 AAZX01000449 UFQT01002153 SSX32787.1 GDHF01028428 JAI23886.1 GAIX01000972 JAA91588.1 NNAY01001281 OXU24506.1 AJWK01005120 AXCM01000651 GBYB01008886 JAG78653.1 GFDF01009278 JAV04806.1 GFDF01009164 JAV04920.1 KQ434948 KZC12473.1 GAMC01002554 JAC04002.1 JRES01000007 KNC34936.1 GFDL01010291 JAV24754.1 KQ435710 KOX79925.1 DS232065 EDS33686.1 CH477443 EAT40804.1 CP012526 ALC46209.1 CH964232 EDW80765.1 CH940652 EDW58978.1 GDAI01000913 JAI16690.1 KZ288271 PBC29780.1 GANO01002136 JAB57735.1 KK852996 KDR12701.1 JXUM01023446 KQ560661 KXJ81388.1 OUUW01000007 SPP82672.1 CH902617 EDV43138.1 CM000070 EAL27982.1 AXCN02000218 JXUM01054531 KQ561824 KXJ77423.1 CH933806 EDW14295.1 CM000160 EDW96497.1 AE014297 BT003501 CH954181 EDV49945.1 CH480815 EDW43052.1 ATLV01019399 KE525269 KFB44134.1 GL447768 EFN85987.1 AAAB01008859 EAA08101.2 APCN01000421 CH916369 EDV92718.1 EZ424016 ADD20292.1 CVRI01000026 CRK92354.1 GEDC01003606 JAS33692.1 GECU01002722 JAT04985.1 GGFJ01007234 MBW56375.1 GGFJ01007232 MBW56373.1 GGFJ01007233 MBW56374.1 ADMH02001252 ETN63460.1 GECZ01023053 JAS46716.1 KQ978231 KYM96131.1 JXJN01018364 CCAG010010901 ADTU01020821 KK107432 QOIP01000001 EZA50901.1 RLU27501.1 GL444634 EFN60768.1 GEBQ01018022 JAT21955.1 APGK01058833 KB741292 KB632334 ENN70314.1 ERL92758.1 KQ981940 KYN32715.1 BT127070 AEE62032.1 GEZM01015649 GEZM01015648 JAV91492.1 GL888334 EGI62558.1 KQ976529 KYM81753.1 KQ983115 KYQ47286.1 KQ980745 KYN13656.1 ABLF02037531 KQ971371 EFA10140.1 GGMS01002964 MBY72167.1 GBHO01006851 GBRD01004034 GDHC01007077 JAG36753.1 JAG61787.1 JAQ11552.1
KQ459606 KPI91115.1 KQ460947 KPJ10522.1 GDQN01003021 JAT88033.1 BABH01001346 UFQS01001768 UFQT01001768 SSX12281.1 SSX31732.1 AAZX01000449 UFQT01002153 SSX32787.1 GDHF01028428 JAI23886.1 GAIX01000972 JAA91588.1 NNAY01001281 OXU24506.1 AJWK01005120 AXCM01000651 GBYB01008886 JAG78653.1 GFDF01009278 JAV04806.1 GFDF01009164 JAV04920.1 KQ434948 KZC12473.1 GAMC01002554 JAC04002.1 JRES01000007 KNC34936.1 GFDL01010291 JAV24754.1 KQ435710 KOX79925.1 DS232065 EDS33686.1 CH477443 EAT40804.1 CP012526 ALC46209.1 CH964232 EDW80765.1 CH940652 EDW58978.1 GDAI01000913 JAI16690.1 KZ288271 PBC29780.1 GANO01002136 JAB57735.1 KK852996 KDR12701.1 JXUM01023446 KQ560661 KXJ81388.1 OUUW01000007 SPP82672.1 CH902617 EDV43138.1 CM000070 EAL27982.1 AXCN02000218 JXUM01054531 KQ561824 KXJ77423.1 CH933806 EDW14295.1 CM000160 EDW96497.1 AE014297 BT003501 CH954181 EDV49945.1 CH480815 EDW43052.1 ATLV01019399 KE525269 KFB44134.1 GL447768 EFN85987.1 AAAB01008859 EAA08101.2 APCN01000421 CH916369 EDV92718.1 EZ424016 ADD20292.1 CVRI01000026 CRK92354.1 GEDC01003606 JAS33692.1 GECU01002722 JAT04985.1 GGFJ01007234 MBW56375.1 GGFJ01007232 MBW56373.1 GGFJ01007233 MBW56374.1 ADMH02001252 ETN63460.1 GECZ01023053 JAS46716.1 KQ978231 KYM96131.1 JXJN01018364 CCAG010010901 ADTU01020821 KK107432 QOIP01000001 EZA50901.1 RLU27501.1 GL444634 EFN60768.1 GEBQ01018022 JAT21955.1 APGK01058833 KB741292 KB632334 ENN70314.1 ERL92758.1 KQ981940 KYN32715.1 BT127070 AEE62032.1 GEZM01015649 GEZM01015648 JAV91492.1 GL888334 EGI62558.1 KQ976529 KYM81753.1 KQ983115 KYQ47286.1 KQ980745 KYN13656.1 ABLF02037531 KQ971371 EFA10140.1 GGMS01002964 MBY72167.1 GBHO01006851 GBRD01004034 GDHC01007077 JAG36753.1 JAG61787.1 JAQ11552.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000053240
UP000005204
UP000002358
+ More
UP000215335 UP000092461 UP000075883 UP000075920 UP000095301 UP000076502 UP000005203 UP000037069 UP000095300 UP000053105 UP000002320 UP000008820 UP000092553 UP000007798 UP000008792 UP000242457 UP000027135 UP000069940 UP000249989 UP000268350 UP000007801 UP000192221 UP000001819 UP000075886 UP000009192 UP000002282 UP000000803 UP000008711 UP000001292 UP000030765 UP000091820 UP000075884 UP000075903 UP000075900 UP000075885 UP000069272 UP000078200 UP000075880 UP000075902 UP000076407 UP000008237 UP000075882 UP000007062 UP000075840 UP000001070 UP000183832 UP000092445 UP000075881 UP000000673 UP000092443 UP000078542 UP000092460 UP000092444 UP000005205 UP000053097 UP000279307 UP000000311 UP000019118 UP000030742 UP000078541 UP000079169 UP000075901 UP000007755 UP000078540 UP000075809 UP000078492 UP000192223 UP000007819 UP000076408 UP000007266
UP000215335 UP000092461 UP000075883 UP000075920 UP000095301 UP000076502 UP000005203 UP000037069 UP000095300 UP000053105 UP000002320 UP000008820 UP000092553 UP000007798 UP000008792 UP000242457 UP000027135 UP000069940 UP000249989 UP000268350 UP000007801 UP000192221 UP000001819 UP000075886 UP000009192 UP000002282 UP000000803 UP000008711 UP000001292 UP000030765 UP000091820 UP000075884 UP000075903 UP000075900 UP000075885 UP000069272 UP000078200 UP000075880 UP000075902 UP000076407 UP000008237 UP000075882 UP000007062 UP000075840 UP000001070 UP000183832 UP000092445 UP000075881 UP000000673 UP000092443 UP000078542 UP000092460 UP000092444 UP000005205 UP000053097 UP000279307 UP000000311 UP000019118 UP000030742 UP000078541 UP000079169 UP000075901 UP000007755 UP000078540 UP000075809 UP000078492 UP000192223 UP000007819 UP000076408 UP000007266
Interpro
Gene 3D
ProteinModelPortal
A0A2A4K7Z0
A0A212ESA0
A0A2H1V8F2
A0A194PJ54
A0A194QYI2
A0A1E1WLY0
+ More
H9IUC4 A0A336L574 K7J4T3 A0A336MX09 A0A0K8UBE2 S4PNA2 A0A232F149 A0A1B0CBG2 A0A182MRR5 A0A0C9RP07 A0A1L8DED5 A0A1L8DEZ0 A0A182VX57 A0A1I8MES2 A0A154PKX3 A0A088ASJ3 W8C917 A0A0L0CRU3 A0A1I8PRB0 A0A1Q3FB01 A0A0M9A9E3 A0A1S4FGL2 B0WS63 Q171Y2 A0A0M3QXP1 B4NAF1 B4M5Q2 A0A0K8TRY9 A0A2A3EDK5 U5EJR6 A0A067QUH2 A0A182G1I3 A0A3B0JQL9 B3M0D2 A0A1W4VHG1 Q298H3 A0A182QHB8 A0A182GCP6 B4K7W1 B4PT36 Q9VHT4 B3NZU7 B4HLM5 A0A084W1P0 A0A1A9WY54 A0A182NA86 A0A182VD60 A0A182R3N3 A0A182PA61 A0A182F8D0 A0A1A9UZ23 A0A182IVJ0 A0A182TU73 A0A182XI94 E2BEE2 A0A182LFB2 Q7QC10 A0A182HT81 B4JH27 D3TRG4 A0A1J1HXS6 A0A1A9Z9M3 A0A182JX79 A0A1B6E6Z6 A0A1B6K0M0 A0A2M4BTH8 A0A2M4BTP3 A0A2M4BTN7 W5JKP2 A0A1B6F945 A0A1A9XG71 A0A195C7J3 A0A1B0BQ30 A0A1B0GAV4 A0A158NMV8 A0A026W5L1 E2B0K8 A0A1B6LE31 N6SRR0 A0A195EWR2 J3JV05 A0A1S3DHQ6 A0A1Y1N0R0 A0A182SDQ1 F4WT95 A0A195BB45 A0A151WHH2 A0A151IYP0 A0A1W4XDX3 J9JP93 A0A182YKI2 D6X1P8 A0A2S2Q391 A0A0A9Z075
H9IUC4 A0A336L574 K7J4T3 A0A336MX09 A0A0K8UBE2 S4PNA2 A0A232F149 A0A1B0CBG2 A0A182MRR5 A0A0C9RP07 A0A1L8DED5 A0A1L8DEZ0 A0A182VX57 A0A1I8MES2 A0A154PKX3 A0A088ASJ3 W8C917 A0A0L0CRU3 A0A1I8PRB0 A0A1Q3FB01 A0A0M9A9E3 A0A1S4FGL2 B0WS63 Q171Y2 A0A0M3QXP1 B4NAF1 B4M5Q2 A0A0K8TRY9 A0A2A3EDK5 U5EJR6 A0A067QUH2 A0A182G1I3 A0A3B0JQL9 B3M0D2 A0A1W4VHG1 Q298H3 A0A182QHB8 A0A182GCP6 B4K7W1 B4PT36 Q9VHT4 B3NZU7 B4HLM5 A0A084W1P0 A0A1A9WY54 A0A182NA86 A0A182VD60 A0A182R3N3 A0A182PA61 A0A182F8D0 A0A1A9UZ23 A0A182IVJ0 A0A182TU73 A0A182XI94 E2BEE2 A0A182LFB2 Q7QC10 A0A182HT81 B4JH27 D3TRG4 A0A1J1HXS6 A0A1A9Z9M3 A0A182JX79 A0A1B6E6Z6 A0A1B6K0M0 A0A2M4BTH8 A0A2M4BTP3 A0A2M4BTN7 W5JKP2 A0A1B6F945 A0A1A9XG71 A0A195C7J3 A0A1B0BQ30 A0A1B0GAV4 A0A158NMV8 A0A026W5L1 E2B0K8 A0A1B6LE31 N6SRR0 A0A195EWR2 J3JV05 A0A1S3DHQ6 A0A1Y1N0R0 A0A182SDQ1 F4WT95 A0A195BB45 A0A151WHH2 A0A151IYP0 A0A1W4XDX3 J9JP93 A0A182YKI2 D6X1P8 A0A2S2Q391 A0A0A9Z075
PDB
5OGK
E-value=0.00208662,
Score=96
Ontologies
GO
PANTHER
Topology
Subcellular location
Golgi apparatus membrane
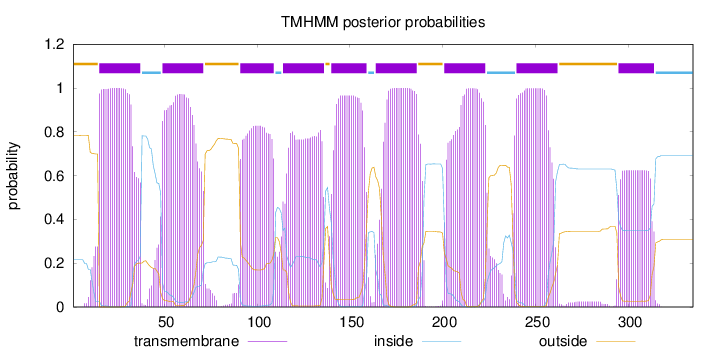
Length:
335
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
174.17752
Exp number, first 60 AAs:
34.86875
Total prob of N-in:
0.21612
POSSIBLE N-term signal
sequence
outside
1 - 14
TMhelix
15 - 37
inside
38 - 48
TMhelix
49 - 71
outside
72 - 90
TMhelix
91 - 109
inside
110 - 113
TMhelix
114 - 136
outside
137 - 139
TMhelix
140 - 159
inside
160 - 163
TMhelix
164 - 186
outside
187 - 200
TMhelix
201 - 223
inside
224 - 239
TMhelix
240 - 262
outside
263 - 294
TMhelix
295 - 314
inside
315 - 335
Population Genetic Test Statistics
Pi
162.250424
Theta
148.479907
Tajima's D
0.180905
CLR
0.617753
CSRT
0.420778961051947
Interpretation
Uncertain
