Gene
KWMTBOMO07655 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001292
Annotation
progesterone_receptor_membrane_component_2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.074
Sequence
CDS
ATGGATTCACCACCAGAGGTTGCTCCCTTGAAAAAACTACCTAAATTGCGAAAAGATATGACCGCAGTAGAACTACGGCAGTACGACGGGACACAGGAGGGTGGCAGAGTGTTGATGGCTGTGAATGGGTGGATATTCGATGTGACTAGAGGGAATCGGTTCTACGGTCCCGGTGGGCCATATGCTGTTTTTGGAGGAAGAGATGCCACAAGAGGACTCGCCACGTTCTCTGTCACAGCACCTGAAAAGGACTATGATGACTTGAGTGATCTCAACTCTATGGAAATGGAATCAGTGAGGGAATGGGAGGAACAATTTAGAGAGAATTATGATCTGGTCGGTCGACTTCTGAGGCCTGGTGAAGAACCCAGGAACTACTCGGACGAAGAACCCGAAGAAGCAGACACGTCCGCTCTCCCCAACGACGACAAGAAAACAAATTGA
Protein
MDSPPEVAPLKKLPKLRKDMTAVELRQYDGTQEGGRVLMAVNGWIFDVTRGNRFYGPGGPYAVFGGRDATRGLATFSVTAPEKDYDDLSDLNSMEMESVREWEEQFRENYDLVGRLLRPGEEPRNYSDEEPEEADTSALPNDDKKTN
Summary
Similarity
Belongs to the cytochrome b5 family.
Uniprot
Q2F5N3
H9IVL2
A0A2A4K771
A0A1E1WG86
S4PPK0
A0A3S2N587
+ More
A0A194PEJ6 A0A194R3V4 A0A1Z2RRL1 A0A212ES84 D2A0J9 A0A0K8TN47 V5GVR3 A0A1J1ITA0 U5EFV8 N6TFW3 J3JTH6 A0A1Y1KZN4 A0A084VV69 A0A182JCQ5 E0VCF2 A0A146LKZ3 A0A232FAV2 A0A2A4IU98 K7ISI4 Q9VXM4 B4IKF4 A0A1L8E169 A0A2M4C072 T1E728 A0A2M3YZT1 A0A2M4A4S7 A0A2M4A9P7 A0A161MQT9 I4DMH8 A0A023F8C5 A0A182F602 T1P889 A0A2P8YWL4 B3NXI0 B4PX88 A0A1L8E124 W5J1G2 A0A023FUB9 A0A023GG41 G3MM04 L7M742 A0A1E1XUF4 A0A023FFN3 A0A023EJT3 A0A069DP82 A0A224XQ28 A0A0C9SD70 A0A158P164 A0A151HZI4 T1IQC1 A0A151J3M1 A0A131XCD8 E2AQY5 A0A195CK83 I4DJ14 A0A1L8EE16 A0A026W1I3 A0A0N7Z989 A0A1L8EE88 A0A194QZ24 A0A1I8PPN9 A0A154PIQ9 A0A087TDQ7 A0A195F3W5 R4G4F6 A0A151X8Z2 A0A2H1V8I6 A0A194PJ68 A0A1E1WBG9 A0A1A9WA56 E2BWH9 A0A0J7NNW8 F4W9A3 A0A293MV77 A0A1A9UM60 A0A2R5LDV5 B0WV18 A0A1A9Z8I9 A0A3G1T1I5 A0A1A9XXZ9 A0A1B0BX16 A0A3S2LW93 S4PBX3 A0A2P6L8S6 A0A131YZ06 E9IKT2 A0A1E1XBI1 E0X658 T1IHL6 A0A023EKZ7 B4KIH4 A0A2R7WYQ7 A0A023EL38 A0A023ENE9
A0A194PEJ6 A0A194R3V4 A0A1Z2RRL1 A0A212ES84 D2A0J9 A0A0K8TN47 V5GVR3 A0A1J1ITA0 U5EFV8 N6TFW3 J3JTH6 A0A1Y1KZN4 A0A084VV69 A0A182JCQ5 E0VCF2 A0A146LKZ3 A0A232FAV2 A0A2A4IU98 K7ISI4 Q9VXM4 B4IKF4 A0A1L8E169 A0A2M4C072 T1E728 A0A2M3YZT1 A0A2M4A4S7 A0A2M4A9P7 A0A161MQT9 I4DMH8 A0A023F8C5 A0A182F602 T1P889 A0A2P8YWL4 B3NXI0 B4PX88 A0A1L8E124 W5J1G2 A0A023FUB9 A0A023GG41 G3MM04 L7M742 A0A1E1XUF4 A0A023FFN3 A0A023EJT3 A0A069DP82 A0A224XQ28 A0A0C9SD70 A0A158P164 A0A151HZI4 T1IQC1 A0A151J3M1 A0A131XCD8 E2AQY5 A0A195CK83 I4DJ14 A0A1L8EE16 A0A026W1I3 A0A0N7Z989 A0A1L8EE88 A0A194QZ24 A0A1I8PPN9 A0A154PIQ9 A0A087TDQ7 A0A195F3W5 R4G4F6 A0A151X8Z2 A0A2H1V8I6 A0A194PJ68 A0A1E1WBG9 A0A1A9WA56 E2BWH9 A0A0J7NNW8 F4W9A3 A0A293MV77 A0A1A9UM60 A0A2R5LDV5 B0WV18 A0A1A9Z8I9 A0A3G1T1I5 A0A1A9XXZ9 A0A1B0BX16 A0A3S2LW93 S4PBX3 A0A2P6L8S6 A0A131YZ06 E9IKT2 A0A1E1XBI1 E0X658 T1IHL6 A0A023EKZ7 B4KIH4 A0A2R7WYQ7 A0A023EL38 A0A023ENE9
Pubmed
19121390
23622113
26354079
28605547
22118469
18362917
+ More
19820115 26369729 23537049 22516182 28004739 24438588 20566863 26823975 28648823 20075255 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 22651552 25474469 25315136 29403074 18057021 17550304 20920257 23761445 22216098 25576852 29209593 24945155 26334808 26131772 21347285 28049606 20798317 24508170 30249741 27129103 21719571 26830274 21282665 28503490 20566361
19820115 26369729 23537049 22516182 28004739 24438588 20566863 26823975 28648823 20075255 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 22651552 25474469 25315136 29403074 18057021 17550304 20920257 23761445 22216098 25576852 29209593 24945155 26334808 26131772 21347285 28049606 20798317 24508170 30249741 27129103 21719571 26830274 21282665 28503490 20566361
EMBL
DQ311390
ABD36334.1
BABH01001348
BABH01001349
NWSH01000056
PCG80125.1
+ More
GDQN01005173 JAT85881.1 GAIX01000207 JAA92353.1 RSAL01000408 RVE41871.1 KQ459606 KPI91114.1 KQ460947 KPJ10521.1 KY963167 ASA46487.1 AGBW02012839 OWR44366.1 KQ971338 EFA02516.1 GDAI01002253 JAI15350.1 GALX01004133 JAB64333.1 CVRI01000057 CRL02334.1 GANO01003716 JAB56155.1 APGK01029308 APGK01029309 KB740732 ENN79274.1 BT126533 KB632390 AEE61497.1 ERL94417.1 GEZM01072038 JAV65620.1 ATLV01017157 KE525157 KFB41863.1 DS235053 EEB11058.1 GDHC01009886 JAQ08743.1 NNAY01000581 OXU27563.1 NWSH01008064 PCG62710.1 AE014298 AY061163 AB290904 AB290905 AAF48534.1 AAL28711.1 ACZ95300.1 AGB95429.1 BAF95203.1 CH480852 EDW51546.1 GFDF01001655 JAV12429.1 GGFJ01009576 MBW58717.1 GAMD01003402 JAA98188.1 GGFM01001022 MBW21773.1 GGFK01002401 MBW35722.1 GGFK01004160 MBW37481.1 GEMB01002120 JAS01059.1 AK402496 BAM19118.1 GBBI01001167 JAC17545.1 KA644812 AFP59441.1 PYGN01000316 PSN48647.1 CH954180 EDV46939.1 KQS30285.1 KQS30286.1 KQS30287.1 CM000162 EDX01851.1 KRK06369.1 KRK06370.1 GFDF01001656 JAV12428.1 ADMH02002182 ETN58007.1 GBBL01001909 JAC25411.1 GBBM01002554 JAC32864.1 JO842905 AEO34522.1 GACK01006100 JAA58934.1 GFAA01000496 JAU02939.1 GBBK01003696 JAC20786.1 GAPW01004262 JAC09336.1 GBGD01003041 JAC85848.1 GFTR01003242 JAW13184.1 GBZX01001997 JAG90743.1 ADTU01001047 KQ976664 KYM78296.1 JH431303 KQ980275 KYN17021.1 GEFH01003257 JAP65324.1 GL441846 EFN64131.1 KQ977720 KYN00484.1 AK401282 BAM17904.1 GFDG01001847 JAV16952.1 KK107488 QOIP01000012 EZA49922.1 RLU16260.1 GDKW01001219 JAI55376.1 GFDG01001845 JAV16954.1 KPJ10534.1 KQ434926 KZC11739.1 KK114763 KFM63246.1 KQ981855 KYN34867.1 GAHY01001405 JAA76105.1 KQ982402 KYQ56847.1 ODYU01001210 SOQ37116.1 KPI91130.1 GDQN01010066 GDQN01006731 GDQN01001056 GDQN01000301 JAT80988.1 JAT84323.1 JAT89998.1 JAT90753.1 GL451131 EFN79935.1 LBMM01002911 KMQ94195.1 GL888002 EGI69285.1 GFWV01019291 MAA44019.1 GGLE01003479 MBY07605.1 DS232115 EDS35346.1 MG992419 AXY94857.1 JXJN01022027 RSAL01000161 RVE45403.1 GAIX01008115 JAA84445.1 MWRG01000919 PRD34990.1 GEDV01004144 JAP84413.1 GL764026 EFZ18849.1 GFAC01002630 JAT96558.1 GQ505293 ADK38544.1 JH429930 GAPW01003741 JAC09857.1 CH933807 EDW13471.1 KK856015 PTY24642.1 GAPW01003670 JAC09928.1 GAPW01003669 JAC09929.1
GDQN01005173 JAT85881.1 GAIX01000207 JAA92353.1 RSAL01000408 RVE41871.1 KQ459606 KPI91114.1 KQ460947 KPJ10521.1 KY963167 ASA46487.1 AGBW02012839 OWR44366.1 KQ971338 EFA02516.1 GDAI01002253 JAI15350.1 GALX01004133 JAB64333.1 CVRI01000057 CRL02334.1 GANO01003716 JAB56155.1 APGK01029308 APGK01029309 KB740732 ENN79274.1 BT126533 KB632390 AEE61497.1 ERL94417.1 GEZM01072038 JAV65620.1 ATLV01017157 KE525157 KFB41863.1 DS235053 EEB11058.1 GDHC01009886 JAQ08743.1 NNAY01000581 OXU27563.1 NWSH01008064 PCG62710.1 AE014298 AY061163 AB290904 AB290905 AAF48534.1 AAL28711.1 ACZ95300.1 AGB95429.1 BAF95203.1 CH480852 EDW51546.1 GFDF01001655 JAV12429.1 GGFJ01009576 MBW58717.1 GAMD01003402 JAA98188.1 GGFM01001022 MBW21773.1 GGFK01002401 MBW35722.1 GGFK01004160 MBW37481.1 GEMB01002120 JAS01059.1 AK402496 BAM19118.1 GBBI01001167 JAC17545.1 KA644812 AFP59441.1 PYGN01000316 PSN48647.1 CH954180 EDV46939.1 KQS30285.1 KQS30286.1 KQS30287.1 CM000162 EDX01851.1 KRK06369.1 KRK06370.1 GFDF01001656 JAV12428.1 ADMH02002182 ETN58007.1 GBBL01001909 JAC25411.1 GBBM01002554 JAC32864.1 JO842905 AEO34522.1 GACK01006100 JAA58934.1 GFAA01000496 JAU02939.1 GBBK01003696 JAC20786.1 GAPW01004262 JAC09336.1 GBGD01003041 JAC85848.1 GFTR01003242 JAW13184.1 GBZX01001997 JAG90743.1 ADTU01001047 KQ976664 KYM78296.1 JH431303 KQ980275 KYN17021.1 GEFH01003257 JAP65324.1 GL441846 EFN64131.1 KQ977720 KYN00484.1 AK401282 BAM17904.1 GFDG01001847 JAV16952.1 KK107488 QOIP01000012 EZA49922.1 RLU16260.1 GDKW01001219 JAI55376.1 GFDG01001845 JAV16954.1 KPJ10534.1 KQ434926 KZC11739.1 KK114763 KFM63246.1 KQ981855 KYN34867.1 GAHY01001405 JAA76105.1 KQ982402 KYQ56847.1 ODYU01001210 SOQ37116.1 KPI91130.1 GDQN01010066 GDQN01006731 GDQN01001056 GDQN01000301 JAT80988.1 JAT84323.1 JAT89998.1 JAT90753.1 GL451131 EFN79935.1 LBMM01002911 KMQ94195.1 GL888002 EGI69285.1 GFWV01019291 MAA44019.1 GGLE01003479 MBY07605.1 DS232115 EDS35346.1 MG992419 AXY94857.1 JXJN01022027 RSAL01000161 RVE45403.1 GAIX01008115 JAA84445.1 MWRG01000919 PRD34990.1 GEDV01004144 JAP84413.1 GL764026 EFZ18849.1 GFAC01002630 JAT96558.1 GQ505293 ADK38544.1 JH429930 GAPW01003741 JAC09857.1 CH933807 EDW13471.1 KK856015 PTY24642.1 GAPW01003670 JAC09928.1 GAPW01003669 JAC09929.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053240
UP000007151
+ More
UP000007266 UP000183832 UP000019118 UP000030742 UP000030765 UP000075880 UP000009046 UP000215335 UP000002358 UP000000803 UP000001292 UP000069272 UP000095301 UP000245037 UP000008711 UP000002282 UP000000673 UP000005205 UP000078540 UP000078492 UP000000311 UP000078542 UP000053097 UP000279307 UP000095300 UP000076502 UP000054359 UP000078541 UP000075809 UP000091820 UP000008237 UP000036403 UP000007755 UP000078200 UP000002320 UP000092445 UP000092443 UP000092460 UP000009192
UP000007266 UP000183832 UP000019118 UP000030742 UP000030765 UP000075880 UP000009046 UP000215335 UP000002358 UP000000803 UP000001292 UP000069272 UP000095301 UP000245037 UP000008711 UP000002282 UP000000673 UP000005205 UP000078540 UP000078492 UP000000311 UP000078542 UP000053097 UP000279307 UP000095300 UP000076502 UP000054359 UP000078541 UP000075809 UP000091820 UP000008237 UP000036403 UP000007755 UP000078200 UP000002320 UP000092445 UP000092443 UP000092460 UP000009192
Pfam
PF00173 Cyt-b5
SUPFAM
SSF55856
SSF55856
Gene 3D
ProteinModelPortal
Q2F5N3
H9IVL2
A0A2A4K771
A0A1E1WG86
S4PPK0
A0A3S2N587
+ More
A0A194PEJ6 A0A194R3V4 A0A1Z2RRL1 A0A212ES84 D2A0J9 A0A0K8TN47 V5GVR3 A0A1J1ITA0 U5EFV8 N6TFW3 J3JTH6 A0A1Y1KZN4 A0A084VV69 A0A182JCQ5 E0VCF2 A0A146LKZ3 A0A232FAV2 A0A2A4IU98 K7ISI4 Q9VXM4 B4IKF4 A0A1L8E169 A0A2M4C072 T1E728 A0A2M3YZT1 A0A2M4A4S7 A0A2M4A9P7 A0A161MQT9 I4DMH8 A0A023F8C5 A0A182F602 T1P889 A0A2P8YWL4 B3NXI0 B4PX88 A0A1L8E124 W5J1G2 A0A023FUB9 A0A023GG41 G3MM04 L7M742 A0A1E1XUF4 A0A023FFN3 A0A023EJT3 A0A069DP82 A0A224XQ28 A0A0C9SD70 A0A158P164 A0A151HZI4 T1IQC1 A0A151J3M1 A0A131XCD8 E2AQY5 A0A195CK83 I4DJ14 A0A1L8EE16 A0A026W1I3 A0A0N7Z989 A0A1L8EE88 A0A194QZ24 A0A1I8PPN9 A0A154PIQ9 A0A087TDQ7 A0A195F3W5 R4G4F6 A0A151X8Z2 A0A2H1V8I6 A0A194PJ68 A0A1E1WBG9 A0A1A9WA56 E2BWH9 A0A0J7NNW8 F4W9A3 A0A293MV77 A0A1A9UM60 A0A2R5LDV5 B0WV18 A0A1A9Z8I9 A0A3G1T1I5 A0A1A9XXZ9 A0A1B0BX16 A0A3S2LW93 S4PBX3 A0A2P6L8S6 A0A131YZ06 E9IKT2 A0A1E1XBI1 E0X658 T1IHL6 A0A023EKZ7 B4KIH4 A0A2R7WYQ7 A0A023EL38 A0A023ENE9
A0A194PEJ6 A0A194R3V4 A0A1Z2RRL1 A0A212ES84 D2A0J9 A0A0K8TN47 V5GVR3 A0A1J1ITA0 U5EFV8 N6TFW3 J3JTH6 A0A1Y1KZN4 A0A084VV69 A0A182JCQ5 E0VCF2 A0A146LKZ3 A0A232FAV2 A0A2A4IU98 K7ISI4 Q9VXM4 B4IKF4 A0A1L8E169 A0A2M4C072 T1E728 A0A2M3YZT1 A0A2M4A4S7 A0A2M4A9P7 A0A161MQT9 I4DMH8 A0A023F8C5 A0A182F602 T1P889 A0A2P8YWL4 B3NXI0 B4PX88 A0A1L8E124 W5J1G2 A0A023FUB9 A0A023GG41 G3MM04 L7M742 A0A1E1XUF4 A0A023FFN3 A0A023EJT3 A0A069DP82 A0A224XQ28 A0A0C9SD70 A0A158P164 A0A151HZI4 T1IQC1 A0A151J3M1 A0A131XCD8 E2AQY5 A0A195CK83 I4DJ14 A0A1L8EE16 A0A026W1I3 A0A0N7Z989 A0A1L8EE88 A0A194QZ24 A0A1I8PPN9 A0A154PIQ9 A0A087TDQ7 A0A195F3W5 R4G4F6 A0A151X8Z2 A0A2H1V8I6 A0A194PJ68 A0A1E1WBG9 A0A1A9WA56 E2BWH9 A0A0J7NNW8 F4W9A3 A0A293MV77 A0A1A9UM60 A0A2R5LDV5 B0WV18 A0A1A9Z8I9 A0A3G1T1I5 A0A1A9XXZ9 A0A1B0BX16 A0A3S2LW93 S4PBX3 A0A2P6L8S6 A0A131YZ06 E9IKT2 A0A1E1XBI1 E0X658 T1IHL6 A0A023EKZ7 B4KIH4 A0A2R7WYQ7 A0A023EL38 A0A023ENE9
PDB
4X8Y
E-value=6.11229e-29,
Score=311
Ontologies
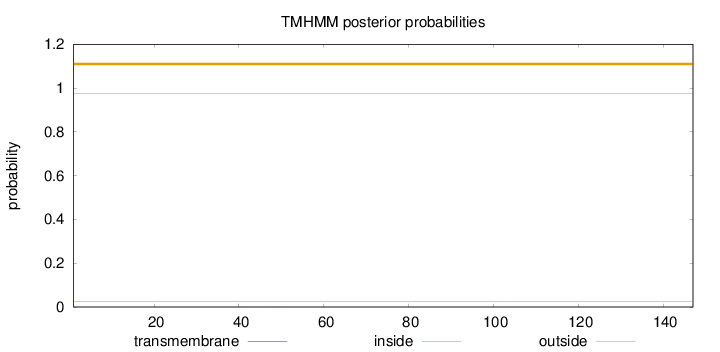
Topology
Length:
147
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00385
Exp number, first 60 AAs:
0.00325
Total prob of N-in:
0.02614
outside
1 - 147
Population Genetic Test Statistics
Pi
244.793384
Theta
177.231766
Tajima's D
0.571375
CLR
0.382203
CSRT
0.532873356332183
Interpretation
Uncertain
