Gene
KWMTBOMO07642
Pre Gene Modal
BGIBMGA000846
Annotation
PREDICTED:_phospholipase_D2_isoform_X3_[Bombyx_mori]
Full name
Phospholipase
Location in the cell
Mitochondrial Reliability : 1.734 Nuclear Reliability : 1.567
Sequence
CDS
ATGGATGTATCCACGCCCACGACCCCGGGCGACTGCCTACCAGGGCTATTCCAGCACATGGACTCCTTGAGCCCAGGGTTGGACTCCGAATTTGACGAAAGTCTTGCAGTACCTGAGTCACTCTCAGTTATAGACATAGATAAAGTTGACAATAAGGATTGTGACGTACTTGCCAAATCGGTGCCATTCAAACACATCCACGAACCACCTATAAAATTCAATTCGGTTCATAGGAAAGTGTTCATTCCTGGAGTCGAAATAAAGGTGAGATTTGTCGAAAACGAAAGGAGTGTTACAACACATCTGCTGAATCCAAATTTGTATACAATATCCCTGCAACATGGCGACTTCACGTGGACGATCAAGAAGCGATACAAGCACATATTAAATCTGCACCAGCAGTTAACACTATATCGGGCGTCGCTAAACATACCGTTTCCAACGAAGGCCCATAAGTCTAGGAGGGCTAGCTTCAAGAACACGGTTGACACTGAAGAGAAAGCCGAGAGGGTGGCTCTCGAAGCTGTCCCGAGGAGTAACTCAAAACGCATCACCAAACCGAGGAAGCGAAGAGGGGCTTTGCCAAGGTTTCCGAAGAAACCAGAAGTCATGATCACGTACGAAGGTATACAATTGCGAATGAAACAATTAGAAGAATATTTATATAATCTCCTGAACATATCCATATACAGGAATCATCACGAAACGGTGAAATTTCTTGAGGTTTCAAACTTATCATTTATATCGGAGCTCGGAAGCAAAGGAAAGGAAGGCATGATACAGAAGCGGACCGGCTCCACGCAGCCCGGTCAAGCAGGATGCAACTGCTTCGGACTCCTGGGTACCGTCGTCTGTGTCAGGTGTAATTACTTCTGCACCGGTCTGGTCTGTGCTAAATGGCAGGAGCGCTGGTTCTTCGTCAAGGACACATTCTTCGGCTACATAAGACCCCGGGACGGAATTGTCAAGGGCATTATGCTGTTTGATCAAGGTTTCGAGGTATCGAGCGGAATGTACTCGACTGGGATGAATCACGGTCTTCAGATACTGAACCAGAGCCGACAAATGGTCATCAAGTGCTGGACGAAACGAAAAAGCAAGGAGTGGATGAACTACCTCAAGACTGTAGCTAATCAGAGTGCCCGCGACTTCACGTACCCGAACGTGCACCACTCGTTCGCGCCCCCCCGGCCCGCCACCCCCGCGCGCGCGCTCGTCGACGGCGCGGAGTACTTCTCGGCGGCGGCCGACGCCATGGAGCTGGCGCGGGAGGAGATCTTCATAGCCGACTGGTGGCTCAGCCCCGAGGTGTACATGAAGCGGCCCGCGCTCAACGGAAACTATTGGAGGCTCGATATGATCTTGAAGAGGAAAGCAGCGCAAGGTGTGAAAATATTTATATTGCTGTATAAAGAAGTTGAAATGGCGCTCGGCATCAACAGCTACTACAGCAAAAGTAGATTAGCTAATGACAATATTAAAGTTTTCCGTCACCCAGACCACGCGAAGGCCGGCGTGTTCTTTTGGGCGCACCACGAGAAGATCGTCGTTGTCGACCAGAGCGTCGCCTTCCTGGGGGGCATCGACCTCTGCTACGGCCGCTGGGACGACCACAGACACAGGTTGACGGATCTGGGTAATATAGCTCAACCGAAGAACTCTATAAGGACTAAGAAGAAGACTTCGAGTTCGCCGGGCGGGGGCGGCCTGTACATACACAACGCGAACGGCATCGACGCCGCGCTGGAGCTGGCCAAGACCTCCAGAGATATCGTCATCGGATTGAATGAGTAA
Protein
MDVSTPTTPGDCLPGLFQHMDSLSPGLDSEFDESLAVPESLSVIDIDKVDNKDCDVLAKSVPFKHIHEPPIKFNSVHRKVFIPGVEIKVRFVENERSVTTHLLNPNLYTISLQHGDFTWTIKKRYKHILNLHQQLTLYRASLNIPFPTKAHKSRRASFKNTVDTEEKAERVALEAVPRSNSKRITKPRKRRGALPRFPKKPEVMITYEGIQLRMKQLEEYLYNLLNISIYRNHHETVKFLEVSNLSFISELGSKGKEGMIQKRTGSTQPGQAGCNCFGLLGTVVCVRCNYFCTGLVCAKWQERWFFVKDTFFGYIRPRDGIVKGIMLFDQGFEVSSGMYSTGMNHGLQILNQSRQMVIKCWTKRKSKEWMNYLKTVANQSARDFTYPNVHHSFAPPRPATPARALVDGAEYFSAAADAMELAREEIFIADWWLSPEVYMKRPALNGNYWRLDMILKRKAAQGVKIFILLYKEVEMALGINSYYSKSRLANDNIKVFRHPDHAKAGVFFWAHHEKIVVVDQSVAFLGGIDLCYGRWDDHRHRLTDLGNIAQPKNSIRTKKKTSSSPGGGGLYIHNANGIDAALELAKTSRDIVIGLNE
Summary
Catalytic Activity
a 1,2-diacyl-sn-glycero-3-phosphocholine + H2O = a 1,2-diacyl-sn-glycero-3-phosphate + choline + H(+)
Similarity
Belongs to the phospholipase D family.
Uniprot
H9IUB6
A0A2A4JXB6
A0A194PEI6
A0A3S2NNW5
A0A2H1VTZ6
A0A034VDU8
+ More
A0A0K8UI57 A0A034VC84 A0A0K8U6Z7 A0A1B0CBQ3 A0A0A1WI48 A0A1L8DQ51 A0A1L8DPX6 A0A1L8DPV0 A0A1I8Q3P5 W8B4F3 W8AQ52 A0A1I8Q3T1 A0A1I8Q3Q2 B4NYT5 B4QD83 A0A0J9R7R1 A0A0R1DN79 B3NB61 A0A3B0J2J4 A0A3B0J2M5 A0A3B0JL11 Q28WX8 A0A3B0IYS8 A0A3B0IZ22 N6WD02 W5JSE9 A0A182FVZ5 B7YZT5 Q9BP34 Q9BP35 Q7KML4 A0A1W4VE50 A0A1W4VEL2 T1PIM7 Q6NR18 A0A2M4ALP0 A0A1I8MGT7 A0A182J218 A0A067R529 A0A182YAC0 B3MJ55 A0A182K565 A0A182LVH5 A0A0N8P0J4 A0A182PN50 A0A182WXZ5 A0A182VHK7 A0A182KUY4 A0A182I615 Q17PR0 A0A1S4H5M9 A0A1Q3FKU9 A0A084WUF4 B4LIM2 A0A1B6IR88 A0A0Q9W3Q0 A0A182RLU9 A0A1B6IY41 Q7PRL1 B4MNV5 A0A182WE79 A0A182SFI3 B0X0U1 A0A182QQM4 A0A1B6DHD6 A0A1B6CGW4 A4UZ54 A0A182NPA5 A0A310SLW0 A0A154NWL7 D2A1R3 A0A139WIU1 A0A195AVS0 A0A224XDL2 F4W630 B4J484 E2B8P7 A0A158P1V7 A0A023EWX2 A0A088AGY1 A0A0C9QG55 A0A195DNV3 A0A195EQD9 A0A336MK21 A0A232FM96 A0A336MPQ6 A0A1J1J2D9 K7ISH9 A0A151I892 A0A151WNY2 A0A1Y1KDD0 A0A1Y1KCY0 A0A1Y1KD64 A0A3L8DYE7
A0A0K8UI57 A0A034VC84 A0A0K8U6Z7 A0A1B0CBQ3 A0A0A1WI48 A0A1L8DQ51 A0A1L8DPX6 A0A1L8DPV0 A0A1I8Q3P5 W8B4F3 W8AQ52 A0A1I8Q3T1 A0A1I8Q3Q2 B4NYT5 B4QD83 A0A0J9R7R1 A0A0R1DN79 B3NB61 A0A3B0J2J4 A0A3B0J2M5 A0A3B0JL11 Q28WX8 A0A3B0IYS8 A0A3B0IZ22 N6WD02 W5JSE9 A0A182FVZ5 B7YZT5 Q9BP34 Q9BP35 Q7KML4 A0A1W4VE50 A0A1W4VEL2 T1PIM7 Q6NR18 A0A2M4ALP0 A0A1I8MGT7 A0A182J218 A0A067R529 A0A182YAC0 B3MJ55 A0A182K565 A0A182LVH5 A0A0N8P0J4 A0A182PN50 A0A182WXZ5 A0A182VHK7 A0A182KUY4 A0A182I615 Q17PR0 A0A1S4H5M9 A0A1Q3FKU9 A0A084WUF4 B4LIM2 A0A1B6IR88 A0A0Q9W3Q0 A0A182RLU9 A0A1B6IY41 Q7PRL1 B4MNV5 A0A182WE79 A0A182SFI3 B0X0U1 A0A182QQM4 A0A1B6DHD6 A0A1B6CGW4 A4UZ54 A0A182NPA5 A0A310SLW0 A0A154NWL7 D2A1R3 A0A139WIU1 A0A195AVS0 A0A224XDL2 F4W630 B4J484 E2B8P7 A0A158P1V7 A0A023EWX2 A0A088AGY1 A0A0C9QG55 A0A195DNV3 A0A195EQD9 A0A336MK21 A0A232FM96 A0A336MPQ6 A0A1J1J2D9 K7ISH9 A0A151I892 A0A151WNY2 A0A1Y1KDD0 A0A1Y1KCY0 A0A1Y1KD64 A0A3L8DYE7
EC Number
3.1.4.4
Pubmed
19121390
26354079
25348373
25830018
24495485
17994087
+ More
17550304 22936249 15632085 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 24845553 25244985 20966253 17510324 12364791 24438588 18362917 19820115 21719571 20798317 21347285 25474469 28648823 20075255 28004739 30249741
17550304 22936249 15632085 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 24845553 25244985 20966253 17510324 12364791 24438588 18362917 19820115 21719571 20798317 21347285 25474469 28648823 20075255 28004739 30249741
EMBL
BABH01001403
BABH01001404
NWSH01000404
PCG76651.1
KQ459606
KPI91104.1
+ More
RSAL01000191 RVE44653.1 ODYU01004430 SOQ44293.1 GAKP01019259 JAC39693.1 GDHF01026269 JAI26045.1 GAKP01019260 JAC39692.1 GDHF01029882 GDHF01029387 GDHF01010539 GDHF01006043 JAI22432.1 JAI22927.1 JAI41775.1 JAI46271.1 AJWK01005749 AJWK01005750 AJWK01005751 GBXI01015578 GBXI01001753 JAC98713.1 JAD12539.1 GFDF01005577 JAV08507.1 GFDF01005578 JAV08506.1 GFDF01005576 JAV08508.1 GAMC01018389 JAB88166.1 GAMC01018388 JAB88167.1 CM000157 EDW89786.2 CM000362 CM002911 EDX05870.1 KMY91723.1 KMY91724.1 KRJ98700.1 CH954177 EDV59826.1 OUUW01000001 SPP73332.1 SPP73333.1 SPP73331.1 CM000071 EAL26538.2 SPP73334.1 SPP73335.1 ENO01964.1 ADMH02000602 ETN65684.1 AE013599 ACL83064.1 AF228315 AAK00728.1 AF228314 AAK00727.1 AF145640 BT046138 AAD38615.2 AAF57264.3 ACI46526.1 KA648626 AFP63255.1 BT010265 AAQ23583.1 GGFK01008207 MBW41528.1 KK852964 KDR13167.1 CH902619 EDV38149.1 AXCM01013130 KPU77292.1 APCN01003547 CH477190 EAT48691.1 AAAB01008848 GFDL01006814 JAV28231.1 ATLV01027083 KE525423 KFB53848.1 CH940648 EDW60392.1 GECU01018286 JAS89420.1 KRF79398.1 GECU01015848 JAS91858.1 EAA07035.3 CH963848 EDW73794.1 DS232245 EDS38333.1 AXCN02001814 GEDC01012220 JAS25078.1 GEDC01029413 GEDC01024630 JAS07885.1 JAS12668.1 AAM68352.3 KQ759902 OAD62011.1 KQ434775 KZC04037.1 KQ971338 EFA02113.2 KYB27775.1 KQ976731 KYM76338.1 GFTR01008548 JAW07878.1 GL887707 EGI70165.1 CH916367 EDW00564.1 GL446361 EFN87953.1 ADTU01006895 GBBI01005080 JAC13632.1 GBYB01002414 JAG72181.1 KQ980724 KYN14164.1 KQ982021 KYN30428.1 UFQT01001485 SSX30754.1 NNAY01000041 OXU31640.1 SSX30753.1 CVRI01000064 CRL05009.1 KQ978397 KYM94197.1 KQ982892 KYQ49612.1 GEZM01086459 JAV59394.1 GEZM01086484 JAV59329.1 GEZM01086486 JAV59324.1 QOIP01000002 RLU25490.1
RSAL01000191 RVE44653.1 ODYU01004430 SOQ44293.1 GAKP01019259 JAC39693.1 GDHF01026269 JAI26045.1 GAKP01019260 JAC39692.1 GDHF01029882 GDHF01029387 GDHF01010539 GDHF01006043 JAI22432.1 JAI22927.1 JAI41775.1 JAI46271.1 AJWK01005749 AJWK01005750 AJWK01005751 GBXI01015578 GBXI01001753 JAC98713.1 JAD12539.1 GFDF01005577 JAV08507.1 GFDF01005578 JAV08506.1 GFDF01005576 JAV08508.1 GAMC01018389 JAB88166.1 GAMC01018388 JAB88167.1 CM000157 EDW89786.2 CM000362 CM002911 EDX05870.1 KMY91723.1 KMY91724.1 KRJ98700.1 CH954177 EDV59826.1 OUUW01000001 SPP73332.1 SPP73333.1 SPP73331.1 CM000071 EAL26538.2 SPP73334.1 SPP73335.1 ENO01964.1 ADMH02000602 ETN65684.1 AE013599 ACL83064.1 AF228315 AAK00728.1 AF228314 AAK00727.1 AF145640 BT046138 AAD38615.2 AAF57264.3 ACI46526.1 KA648626 AFP63255.1 BT010265 AAQ23583.1 GGFK01008207 MBW41528.1 KK852964 KDR13167.1 CH902619 EDV38149.1 AXCM01013130 KPU77292.1 APCN01003547 CH477190 EAT48691.1 AAAB01008848 GFDL01006814 JAV28231.1 ATLV01027083 KE525423 KFB53848.1 CH940648 EDW60392.1 GECU01018286 JAS89420.1 KRF79398.1 GECU01015848 JAS91858.1 EAA07035.3 CH963848 EDW73794.1 DS232245 EDS38333.1 AXCN02001814 GEDC01012220 JAS25078.1 GEDC01029413 GEDC01024630 JAS07885.1 JAS12668.1 AAM68352.3 KQ759902 OAD62011.1 KQ434775 KZC04037.1 KQ971338 EFA02113.2 KYB27775.1 KQ976731 KYM76338.1 GFTR01008548 JAW07878.1 GL887707 EGI70165.1 CH916367 EDW00564.1 GL446361 EFN87953.1 ADTU01006895 GBBI01005080 JAC13632.1 GBYB01002414 JAG72181.1 KQ980724 KYN14164.1 KQ982021 KYN30428.1 UFQT01001485 SSX30754.1 NNAY01000041 OXU31640.1 SSX30753.1 CVRI01000064 CRL05009.1 KQ978397 KYM94197.1 KQ982892 KYQ49612.1 GEZM01086459 JAV59394.1 GEZM01086484 JAV59329.1 GEZM01086486 JAV59324.1 QOIP01000002 RLU25490.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000283053
UP000092461
UP000095300
+ More
UP000002282 UP000000304 UP000008711 UP000268350 UP000001819 UP000000673 UP000069272 UP000000803 UP000192221 UP000095301 UP000075880 UP000027135 UP000076408 UP000007801 UP000075881 UP000075883 UP000075885 UP000076407 UP000075903 UP000075882 UP000075840 UP000008820 UP000030765 UP000008792 UP000075900 UP000007062 UP000007798 UP000075920 UP000075901 UP000002320 UP000075886 UP000075884 UP000076502 UP000007266 UP000078540 UP000007755 UP000001070 UP000008237 UP000005205 UP000005203 UP000078492 UP000078541 UP000215335 UP000183832 UP000002358 UP000078542 UP000075809 UP000279307
UP000002282 UP000000304 UP000008711 UP000268350 UP000001819 UP000000673 UP000069272 UP000000803 UP000192221 UP000095301 UP000075880 UP000027135 UP000076408 UP000007801 UP000075881 UP000075883 UP000075885 UP000076407 UP000075903 UP000075882 UP000075840 UP000008820 UP000030765 UP000008792 UP000075900 UP000007062 UP000007798 UP000075920 UP000075901 UP000002320 UP000075886 UP000075884 UP000076502 UP000007266 UP000078540 UP000007755 UP000001070 UP000008237 UP000005205 UP000005203 UP000078492 UP000078541 UP000215335 UP000183832 UP000002358 UP000078542 UP000075809 UP000279307
Interpro
SUPFAM
SSF64268
SSF64268
Gene 3D
ProteinModelPortal
H9IUB6
A0A2A4JXB6
A0A194PEI6
A0A3S2NNW5
A0A2H1VTZ6
A0A034VDU8
+ More
A0A0K8UI57 A0A034VC84 A0A0K8U6Z7 A0A1B0CBQ3 A0A0A1WI48 A0A1L8DQ51 A0A1L8DPX6 A0A1L8DPV0 A0A1I8Q3P5 W8B4F3 W8AQ52 A0A1I8Q3T1 A0A1I8Q3Q2 B4NYT5 B4QD83 A0A0J9R7R1 A0A0R1DN79 B3NB61 A0A3B0J2J4 A0A3B0J2M5 A0A3B0JL11 Q28WX8 A0A3B0IYS8 A0A3B0IZ22 N6WD02 W5JSE9 A0A182FVZ5 B7YZT5 Q9BP34 Q9BP35 Q7KML4 A0A1W4VE50 A0A1W4VEL2 T1PIM7 Q6NR18 A0A2M4ALP0 A0A1I8MGT7 A0A182J218 A0A067R529 A0A182YAC0 B3MJ55 A0A182K565 A0A182LVH5 A0A0N8P0J4 A0A182PN50 A0A182WXZ5 A0A182VHK7 A0A182KUY4 A0A182I615 Q17PR0 A0A1S4H5M9 A0A1Q3FKU9 A0A084WUF4 B4LIM2 A0A1B6IR88 A0A0Q9W3Q0 A0A182RLU9 A0A1B6IY41 Q7PRL1 B4MNV5 A0A182WE79 A0A182SFI3 B0X0U1 A0A182QQM4 A0A1B6DHD6 A0A1B6CGW4 A4UZ54 A0A182NPA5 A0A310SLW0 A0A154NWL7 D2A1R3 A0A139WIU1 A0A195AVS0 A0A224XDL2 F4W630 B4J484 E2B8P7 A0A158P1V7 A0A023EWX2 A0A088AGY1 A0A0C9QG55 A0A195DNV3 A0A195EQD9 A0A336MK21 A0A232FM96 A0A336MPQ6 A0A1J1J2D9 K7ISH9 A0A151I892 A0A151WNY2 A0A1Y1KDD0 A0A1Y1KCY0 A0A1Y1KD64 A0A3L8DYE7
A0A0K8UI57 A0A034VC84 A0A0K8U6Z7 A0A1B0CBQ3 A0A0A1WI48 A0A1L8DQ51 A0A1L8DPX6 A0A1L8DPV0 A0A1I8Q3P5 W8B4F3 W8AQ52 A0A1I8Q3T1 A0A1I8Q3Q2 B4NYT5 B4QD83 A0A0J9R7R1 A0A0R1DN79 B3NB61 A0A3B0J2J4 A0A3B0J2M5 A0A3B0JL11 Q28WX8 A0A3B0IYS8 A0A3B0IZ22 N6WD02 W5JSE9 A0A182FVZ5 B7YZT5 Q9BP34 Q9BP35 Q7KML4 A0A1W4VE50 A0A1W4VEL2 T1PIM7 Q6NR18 A0A2M4ALP0 A0A1I8MGT7 A0A182J218 A0A067R529 A0A182YAC0 B3MJ55 A0A182K565 A0A182LVH5 A0A0N8P0J4 A0A182PN50 A0A182WXZ5 A0A182VHK7 A0A182KUY4 A0A182I615 Q17PR0 A0A1S4H5M9 A0A1Q3FKU9 A0A084WUF4 B4LIM2 A0A1B6IR88 A0A0Q9W3Q0 A0A182RLU9 A0A1B6IY41 Q7PRL1 B4MNV5 A0A182WE79 A0A182SFI3 B0X0U1 A0A182QQM4 A0A1B6DHD6 A0A1B6CGW4 A4UZ54 A0A182NPA5 A0A310SLW0 A0A154NWL7 D2A1R3 A0A139WIU1 A0A195AVS0 A0A224XDL2 F4W630 B4J484 E2B8P7 A0A158P1V7 A0A023EWX2 A0A088AGY1 A0A0C9QG55 A0A195DNV3 A0A195EQD9 A0A336MK21 A0A232FM96 A0A336MPQ6 A0A1J1J2D9 K7ISH9 A0A151I892 A0A151WNY2 A0A1Y1KDD0 A0A1Y1KCY0 A0A1Y1KD64 A0A3L8DYE7
Ontologies
PATHWAY
GO
PANTHER
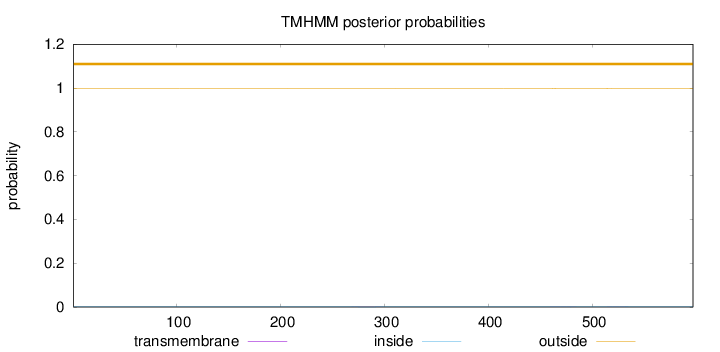
Topology
Length:
597
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0419300000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00070
outside
1 - 597
Population Genetic Test Statistics
Pi
223.658678
Theta
178.155666
Tajima's D
0.865306
CLR
0.19299
CSRT
0.624518774061297
Interpretation
Uncertain
