Gene
KWMTBOMO07638
Pre Gene Modal
BGIBMGA001296
Annotation
PREDICTED:_DNA-binding_protein_D-ETS-4_isoform_X2_[Amyelois_transitella]
Full name
DNA-binding protein D-ETS-4
Transcription factor
Location in the cell
Nuclear Reliability : 4.781
Sequence
CDS
ATGCCACAAACCGTGCCGCGGTCCGGGTGCAGCCCGCCTTCAGCATCACAAGGCGAGAGAGCTGTCCCCACGTCTCCAGCTGATCTACAGCACCTGCTCCGTTTATTAGGGGCCTCGTCGCCGCCAGAATCCATGCTCCACTCTCCACCGAACCTACACACAAAACCTCCGCCACCATATCCAGAGGATAATAACTTCTTGGACAGGCTGTACGATTTTGAAGGATATCCTACCCCGTCTCCTTCGTCAGACGAAGGCTCGATTCCCACGGTGGCGCTGCAACCATCTTCCCCGTACAGTTCACTGCACTACACAACACCAGTGTATATCAAAGAAGAACCAAACAGACTGTCTGTGCCTGGATTTGCCTCACCCTACCCTCTGTCGCCTTCTGGATCCTGTGTATCTTATGGAAGCCATAATCAGTATCCTTCGCCTACACCTCAACCCGAAGAATATATTGACATCGAGCAATTATTAAAAGAGAATCAAATATTACAAGACAGCACACAGCATAGTTACATAACCCCTAAAACAGAGATAGAAGAACCAAGAGATCATATATTATTGAGATCAGTACTCGAAGATACCACATTCCAGAAGAGATTCAATCTTAGACCTGTGCCTTTAGAGTTGGGAAACGTGAAAATGGAGGAGAGCAGTGGTGGTGACGACCTGGTGGCACCGGATATTGATCGTGTGCTGTCGATGGCCATTGAACAATCTAAGAGGGATGTGGATAGCACTTGCACAGTCCTCGGAATATCGCCAGATCCAATGCTTTGGAGTACAGCGGATGTCAAAGCTTGGGTGATGTTTACATTGCAACATTTCAACCAGCCATTAGTACCGTCCGAGTATTTCAATATGGACGGGGCTGCACTGGTGGCTCTTACAGAAGAAGAATTCAACCAGAGAGCTCCTCAGTCGGGAAGCACTCTGTATGCTCAGCTGGAAATATGGAAGGCAGCAAGACACGAGTCGTGGAAGACTTCTCAGTGGACTGACCATCAACCTTCGCCTACACCTGTACCGGCCGCGTTACCCTCAGCCGACGATATGAGTGAAGATGAAGATTCAGAGTCAGCCGGTACCAGTGCGGGAACAACGGTAGGAAAGGCTAAAACTGGCAGCACGCATATACACCTTTGGCAATTCTTGAAAGAGCTTCTAGCTTCGCCCCACATACACGGGTCGGCGATACGATGGCTAGATAGAAGTAACGGTGTCTTCAAGATCGAAGATTCAGTACGCGTCGCGAGGCTCTGGGGCAAGAGAAAGAATAGGCCCGCGATGAACTACGACAAGTTGTCGCGTAGCATCAGGCAGTACTACAAGAAGGGTATTATGAAGAAAACCGAGCGAAGTCAGAGGCTTGTCTACCAGTTCTGCCATCCGTACTGTTTGTGA
Protein
MPQTVPRSGCSPPSASQGERAVPTSPADLQHLLRLLGASSPPESMLHSPPNLHTKPPPPYPEDNNFLDRLYDFEGYPTPSPSSDEGSIPTVALQPSSPYSSLHYTTPVYIKEEPNRLSVPGFASPYPLSPSGSCVSYGSHNQYPSPTPQPEEYIDIEQLLKENQILQDSTQHSYITPKTEIEEPRDHILLRSVLEDTTFQKRFNLRPVPLELGNVKMEESSGGDDLVAPDIDRVLSMAIEQSKRDVDSTCTVLGISPDPMLWSTADVKAWVMFTLQHFNQPLVPSEYFNMDGAALVALTEEEFNQRAPQSGSTLYAQLEIWKAARHESWKTSQWTDHQPSPTPVPAALPSADDMSEDEDSESAGTSAGTTVGKAKTGSTHIHLWQFLKELLASPHIHGSAIRWLDRSNGVFKIEDSVRVARLWGKRKNRPAMNYDKLSRSIRQYYKKGIMKKTERSQRLVYQFCHPYCL
Summary
Description
May have a role in germline development.
Similarity
Belongs to the ETS family.
Keywords
Complete proteome
DNA-binding
Nucleus
Reference proteome
Feature
chain DNA-binding protein D-ETS-4
Uniprot
H9IVL6
A0A2A4JB91
A0A2H1V314
A0A2A4J9D6
A0A2A4J9Q3
A0A2A4JBC0
+ More
A0A2A4JAX8 A0A194PCZ8 A0A0L7KZ20 A0A0N1PJF9 A0A2A4JAA8 A0A1B6FWS9 A0A2J7PQB0 A0A2J7PQB9 A0A1B6FN83 A0A1B6LC17 D2A0N6 A0A0C9QI27 E2ATR9 A0A1B6E6Z2 A0A232FHS1 A0A0M8ZZI6 A0A1Y1JT39 A0A154PEV4 A0A2P8YSM0 N6TJ88 A0A1B0CUK7 A0A1L8DIE6 A0A1L8DI42 A0A0R1EB89 B4PRA4 A0A0B4LHS8 B7FNJ4 P29775 Q29QW5 D5SHS7 A0A1W4VKT9 A0A1W4V8P9 B4IGV9 A0A0P9AT22 B3LX26 B4NAS5 A0A1A9ULY8 A0A1S4F9Z0 Q17AZ6 W8ANS1
A0A2A4JAX8 A0A194PCZ8 A0A0L7KZ20 A0A0N1PJF9 A0A2A4JAA8 A0A1B6FWS9 A0A2J7PQB0 A0A2J7PQB9 A0A1B6FN83 A0A1B6LC17 D2A0N6 A0A0C9QI27 E2ATR9 A0A1B6E6Z2 A0A232FHS1 A0A0M8ZZI6 A0A1Y1JT39 A0A154PEV4 A0A2P8YSM0 N6TJ88 A0A1B0CUK7 A0A1L8DIE6 A0A1L8DI42 A0A0R1EB89 B4PRA4 A0A0B4LHS8 B7FNJ4 P29775 Q29QW5 D5SHS7 A0A1W4VKT9 A0A1W4V8P9 B4IGV9 A0A0P9AT22 B3LX26 B4NAS5 A0A1A9ULY8 A0A1S4F9Z0 Q17AZ6 W8ANS1
Pubmed
EMBL
BABH01001413
BABH01001414
BABH01001415
NWSH01002282
PCG68692.1
ODYU01000237
+ More
SOQ34674.1 PCG68695.1 PCG68691.1 PCG68693.1 PCG68694.1 KQ459606 KPI91102.1 JTDY01004160 KOB68513.1 KQ459836 KPJ19773.1 PCG68696.1 GECZ01015113 JAS54656.1 NEVH01022640 PNF18523.1 PNF18524.1 GECZ01018148 GECZ01011633 JAS51621.1 JAS58136.1 GEBQ01018748 JAT21229.1 KQ971338 EFA02538.2 GBYB01014348 JAG84115.1 GL442691 EFN63152.1 GEDC01003597 JAS33701.1 NNAY01000198 OXU30070.1 KQ435794 KOX74107.1 GEZM01101628 JAV52432.1 KQ434879 KZC09964.1 PYGN01000386 PSN47225.1 APGK01023741 APGK01023742 APGK01023743 APGK01023744 APGK01023745 APGK01023746 APGK01023747 APGK01023748 APGK01023749 KB740512 ENN80509.1 AJWK01029307 AJWK01029308 AJWK01029309 AJWK01029310 GFDF01007947 JAV06137.1 GFDF01007946 JAV06138.1 CM000160 KRK04279.1 KRK04280.1 EDW98468.1 AE014297 AHN57569.1 BT053684 ACK77599.1 M88474 BT024275 ABC86337.1 BT124878 ADG46046.1 CH480837 EDW49077.1 CH902617 KPU80505.1 KPU80506.1 EDV43865.1 CH964232 EDW80889.1 CH477328 EAT43418.1 GAMC01020437 JAB86118.1
SOQ34674.1 PCG68695.1 PCG68691.1 PCG68693.1 PCG68694.1 KQ459606 KPI91102.1 JTDY01004160 KOB68513.1 KQ459836 KPJ19773.1 PCG68696.1 GECZ01015113 JAS54656.1 NEVH01022640 PNF18523.1 PNF18524.1 GECZ01018148 GECZ01011633 JAS51621.1 JAS58136.1 GEBQ01018748 JAT21229.1 KQ971338 EFA02538.2 GBYB01014348 JAG84115.1 GL442691 EFN63152.1 GEDC01003597 JAS33701.1 NNAY01000198 OXU30070.1 KQ435794 KOX74107.1 GEZM01101628 JAV52432.1 KQ434879 KZC09964.1 PYGN01000386 PSN47225.1 APGK01023741 APGK01023742 APGK01023743 APGK01023744 APGK01023745 APGK01023746 APGK01023747 APGK01023748 APGK01023749 KB740512 ENN80509.1 AJWK01029307 AJWK01029308 AJWK01029309 AJWK01029310 GFDF01007947 JAV06137.1 GFDF01007946 JAV06138.1 CM000160 KRK04279.1 KRK04280.1 EDW98468.1 AE014297 AHN57569.1 BT053684 ACK77599.1 M88474 BT024275 ABC86337.1 BT124878 ADG46046.1 CH480837 EDW49077.1 CH902617 KPU80505.1 KPU80506.1 EDV43865.1 CH964232 EDW80889.1 CH477328 EAT43418.1 GAMC01020437 JAB86118.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
H9IVL6
A0A2A4JB91
A0A2H1V314
A0A2A4J9D6
A0A2A4J9Q3
A0A2A4JBC0
+ More
A0A2A4JAX8 A0A194PCZ8 A0A0L7KZ20 A0A0N1PJF9 A0A2A4JAA8 A0A1B6FWS9 A0A2J7PQB0 A0A2J7PQB9 A0A1B6FN83 A0A1B6LC17 D2A0N6 A0A0C9QI27 E2ATR9 A0A1B6E6Z2 A0A232FHS1 A0A0M8ZZI6 A0A1Y1JT39 A0A154PEV4 A0A2P8YSM0 N6TJ88 A0A1B0CUK7 A0A1L8DIE6 A0A1L8DI42 A0A0R1EB89 B4PRA4 A0A0B4LHS8 B7FNJ4 P29775 Q29QW5 D5SHS7 A0A1W4VKT9 A0A1W4V8P9 B4IGV9 A0A0P9AT22 B3LX26 B4NAS5 A0A1A9ULY8 A0A1S4F9Z0 Q17AZ6 W8ANS1
A0A2A4JAX8 A0A194PCZ8 A0A0L7KZ20 A0A0N1PJF9 A0A2A4JAA8 A0A1B6FWS9 A0A2J7PQB0 A0A2J7PQB9 A0A1B6FN83 A0A1B6LC17 D2A0N6 A0A0C9QI27 E2ATR9 A0A1B6E6Z2 A0A232FHS1 A0A0M8ZZI6 A0A1Y1JT39 A0A154PEV4 A0A2P8YSM0 N6TJ88 A0A1B0CUK7 A0A1L8DIE6 A0A1L8DI42 A0A0R1EB89 B4PRA4 A0A0B4LHS8 B7FNJ4 P29775 Q29QW5 D5SHS7 A0A1W4VKT9 A0A1W4V8P9 B4IGV9 A0A0P9AT22 B3LX26 B4NAS5 A0A1A9ULY8 A0A1S4F9Z0 Q17AZ6 W8ANS1
PDB
1YO5
E-value=3.04069e-35,
Score=373
Ontologies
GO
Topology
Subcellular location
Nucleus
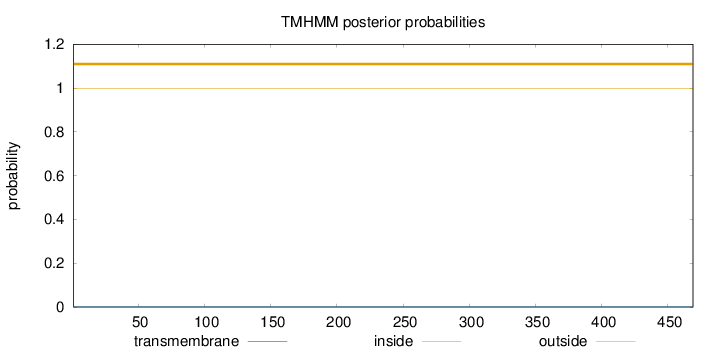
Length:
469
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00021
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00314
outside
1 - 469
Population Genetic Test Statistics
Pi
313.955913
Theta
192.053048
Tajima's D
2.131244
CLR
0.409796
CSRT
0.900154992250388
Interpretation
Uncertain
