Gene
KWMTBOMO07629
Pre Gene Modal
BGIBMGA001298
Annotation
PREDICTED:_MORN_repeat-containing_protein_4_[Amyelois_transitella]
Full name
Translation factor GUF1 homolog, mitochondrial
+ More
MORN repeat-containing protein 4 homolog
MORN repeat-containing protein 4 homolog
Alternative Name
Elongation factor 4 homolog
GTPase GUF1 homolog
Ribosomal back-translocase
Retinophilin
undertaker
GTPase GUF1 homolog
Ribosomal back-translocase
Retinophilin
undertaker
Location in the cell
Cytoplasmic Reliability : 1.316 Mitochondrial Reliability : 1.247
Sequence
CDS
ATGGTCGTGGATCCCGGTGTGGTGAAGACTGGAGCGTATAAATATGAAGACGGCACTCGCTACGTCGGAGAATGGAATGCTAAAGGCCAGAAACATGGGATGGGTCACTTGGTCCTACCCGACGGAACCAGATATGACGGAGCCCTGATCGGAGGATTGTGTTCTGGACTTGGGGTGATGGCGTTTCCTGATGGCGCAAAGTAA
Protein
MVVDPGVVKTGAYKYEDGTRYVGEWNAKGQKHGMGHLVLPDGTRYDGALIGGLCSGLGVMAFPDGAK
Summary
Description
Promotes mitochondrial protein synthesis. May act as a fidelity factor of the translation reaction, by catalyzing a one-codon backward translocation of tRNAs on improperly translocated ribosomes. Binds to mitochondrial ribosomes in a GTP-dependent manner.
Plays a role in promoting axonal degeneration following neuronal injury by toxic insult or trauma (PubMed:22496551). Organizes rhabdomeric components to suppress random activation of the phototransduction cascade and thus increases the signaling fidelity of dark-adapted photoreceptors (PubMed:20107052). The rtp/ninaC complex is required for stability of inad and inac and the normal termination of phototransduction in the retina (PubMed:20739554).
Plays a role in promoting axonal degeneration following neuronal injury by toxic insult or trauma (PubMed:22496551). Organizes rhabdomeric components to suppress random activation of the phototransduction cascade and thus increases the signaling fidelity of dark-adapted photoreceptors (PubMed:20107052). The rtp/ninaC complex is required for stability of inad and inac and the normal termination of phototransduction in the retina (PubMed:20739554).
Catalytic Activity
GTP + H2O = GDP + H(+) + phosphate
Subunit
Interacts with ninaC.
Similarity
Belongs to the GTP-binding elongation factor family. LepA subfamily.
Keywords
Acetylation
Complete proteome
Membrane
Phosphoprotein
Reference proteome
Repeat
Sensory transduction
Vision
Feature
chain MORN repeat-containing protein 4 homolog
Uniprot
H9IVL8
A0A3S2L464
A0A212F3Q2
A0A194QM99
A0A194PDK6
A0A2A3EUI6
+ More
A0A2P8Y895 A0A067RF44 A0A1B0CKH2 A0A182GVT9 A0A1B6FD29 A0A1B6MI08 A0A1J1IIZ5 A0A1B6I651 A0A084VB13 A0A1L8DCG9 T1DQ33 A0A1S3CYE0 Q17PB7 A0A084VB15 A0A182JDS1 A0A182F717 W5JU47 A0A182QRQ6 A0A182HZ81 A0A182V8T2 A0A182TW99 Q7PH89 A0A336LN76 A0A182M6J6 A0A182VS08 A0A310SK63 A0A182NSN3 A0A182WVV7 A0A182RZE1 A0A182KGR3 A0A182P0C9 A0A182T8H4 T1D558 A0A1Q3FRW4 B0WIW1 A0A088AF51 A0A182Y7F9 A0A0N0BEY7 A0A154PST9 A0A0J7KXG4 E2A8P4 A0A3L8D3G6 A0A026WVR0 E9J0W7 A0A151ICZ1 F4WTT2 A0A0K8SNN5 A0A146L576 A0A0A9XWF4 A0A0K8SMT0 E2BRK2 A0A232EZA8 A0A158N9N9 K7JBS3 A0A195BPQ8 B4JSP7 Q296A8 B4GFE9 A0A151XGK6 A0A1I8NX35 B4NI67 A0A034WE33 A0A0M4EQK6 B4KCM3 A0A1A9ZVL4 J9K3X1 A0A1A9Y8I5 A0A1A9VAD9 A0A1B0BLV0 A0A0K8W980 A0A1B0FQT6 B3M170 A0A0A1WJ59 A0A1A9WFS8 B4M4X8 A0A2H8TMD5 A0A2S2NRK1 D2NUK8 A0A1I8NF45 W8AYE1 A0A3B0KFU7 A0A2S2R0P9 B4QW88 B4I3X3 B3P2A4 Q9VN91 A0A1W4UWU5 B4PV68 A0A3M7TA09 E9HAI9 A0A0P4WW95 A0A0P5WN34 A0A0P5H1Y6 A0A0P6CJD3
A0A2P8Y895 A0A067RF44 A0A1B0CKH2 A0A182GVT9 A0A1B6FD29 A0A1B6MI08 A0A1J1IIZ5 A0A1B6I651 A0A084VB13 A0A1L8DCG9 T1DQ33 A0A1S3CYE0 Q17PB7 A0A084VB15 A0A182JDS1 A0A182F717 W5JU47 A0A182QRQ6 A0A182HZ81 A0A182V8T2 A0A182TW99 Q7PH89 A0A336LN76 A0A182M6J6 A0A182VS08 A0A310SK63 A0A182NSN3 A0A182WVV7 A0A182RZE1 A0A182KGR3 A0A182P0C9 A0A182T8H4 T1D558 A0A1Q3FRW4 B0WIW1 A0A088AF51 A0A182Y7F9 A0A0N0BEY7 A0A154PST9 A0A0J7KXG4 E2A8P4 A0A3L8D3G6 A0A026WVR0 E9J0W7 A0A151ICZ1 F4WTT2 A0A0K8SNN5 A0A146L576 A0A0A9XWF4 A0A0K8SMT0 E2BRK2 A0A232EZA8 A0A158N9N9 K7JBS3 A0A195BPQ8 B4JSP7 Q296A8 B4GFE9 A0A151XGK6 A0A1I8NX35 B4NI67 A0A034WE33 A0A0M4EQK6 B4KCM3 A0A1A9ZVL4 J9K3X1 A0A1A9Y8I5 A0A1A9VAD9 A0A1B0BLV0 A0A0K8W980 A0A1B0FQT6 B3M170 A0A0A1WJ59 A0A1A9WFS8 B4M4X8 A0A2H8TMD5 A0A2S2NRK1 D2NUK8 A0A1I8NF45 W8AYE1 A0A3B0KFU7 A0A2S2R0P9 B4QW88 B4I3X3 B3P2A4 Q9VN91 A0A1W4UWU5 B4PV68 A0A3M7TA09 E9HAI9 A0A0P4WW95 A0A0P5WN34 A0A0P5H1Y6 A0A0P6CJD3
EC Number
3.6.5.n1
Pubmed
19121390
22118469
26354079
29403074
24845553
26483478
+ More
24438588 17510324 20920257 23761445 12364791 14747013 17210077 24330624 25244985 20798317 30249741 24508170 21282665 21719571 26823975 25401762 28648823 21347285 20075255 17994087 15632085 25348373 25830018 25315136 24495485 17285308 10731132 12537572 12537569 20107052 20739554 22496551 25822849 17550304 30375419 21292972
24438588 17510324 20920257 23761445 12364791 14747013 17210077 24330624 25244985 20798317 30249741 24508170 21282665 21719571 26823975 25401762 28648823 21347285 20075255 17994087 15632085 25348373 25830018 25315136 24495485 17285308 10731132 12537572 12537569 20107052 20739554 22496551 25822849 17550304 30375419 21292972
EMBL
BABH01001438
RSAL01000167
RVE45248.1
AGBW02010520
OWR48354.1
KQ461198
+ More
KPJ06085.1 KQ459606 KPI91093.1 KZ288185 PBC34876.1 PYGN01000809 PSN40483.1 KK852508 KDR22397.1 AJWK01016269 AJWK01016270 AJWK01016271 AJWK01016272 JXUM01002529 JXUM01002530 JXUM01002531 KQ560140 KXJ84219.1 GECZ01021669 JAS48100.1 GEBQ01004446 JAT35531.1 CVRI01000054 CRL00195.1 GECU01025290 JAS82416.1 ATLV01004846 KE524243 KFB35157.1 GFDF01009921 JAV04163.1 GAMD01002809 JAA98781.1 CH477193 EAT48514.1 ATLV01004847 KFB35159.1 AXCP01002751 ADMH02000081 ETN67872.1 AXCN02000233 APCN01005504 AAAB01008888 EAA44640.4 EGK97065.1 UFQT01000091 SSX19532.1 AXCM01001479 KQ765222 OAD54065.1 GALA01000673 JAA94179.1 GFDL01004757 JAV30288.1 DS231953 EDS28787.1 KQ435821 KOX72291.1 KQ435180 KZC14979.1 LBMM01002304 KMQ95011.1 GL437663 EFN70123.1 QOIP01000014 RLU15047.1 KK107083 EZA60083.1 GL767538 EFZ13487.1 KQ977991 KYM98283.1 GL888344 EGI62397.1 GBRD01011404 GBRD01011403 GBRD01010999 JAG54420.1 GDHC01016332 JAQ02297.1 GBHO01020401 JAG23203.1 GBRD01011400 JAG54424.1 GL449965 EFN81694.1 NNAY01001541 OXU23637.1 ADTU01009725 AAZX01000360 AAZX01011714 AAZX01013565 AAZX01016629 AAZX01025833 KQ976428 KYM87825.1 CH916373 EDV94787.1 CM000070 EAL28550.2 CH479182 EDW34334.1 KQ982169 KYQ59534.1 CH964272 EDW84759.1 GAKP01005141 JAC53811.1 CP012526 ALC46635.1 CH933806 EDW15872.1 ABLF02035568 ABLF02035571 JXJN01016575 GDHF01004677 JAI47637.1 CCAG010014772 CH902617 EDV44340.1 GBXI01015869 JAC98422.1 CH940652 EDW59689.1 GFXV01003522 MBW15327.1 GGMR01007089 MBY19708.1 BT120151 ADB57072.1 GAMC01015415 GAMC01015413 JAB91142.1 OUUW01000008 SPP84616.1 GGMS01014366 MBY83569.1 CM000364 EDX11687.1 CH480821 EDW54916.1 CH954181 EDV47854.1 DQ333317 DQ324736 AE014297 AY060630 CM000160 EDW95737.1 REGN01000043 RNA44944.1 GL732612 EFX71189.1 GDIP01249903 JAI73498.1 GDIP01083969 LRGB01002121 JAM19746.1 KZS09027.1 GDIQ01255989 JAJ95735.1 GDIP01007450 JAM96265.1
KPJ06085.1 KQ459606 KPI91093.1 KZ288185 PBC34876.1 PYGN01000809 PSN40483.1 KK852508 KDR22397.1 AJWK01016269 AJWK01016270 AJWK01016271 AJWK01016272 JXUM01002529 JXUM01002530 JXUM01002531 KQ560140 KXJ84219.1 GECZ01021669 JAS48100.1 GEBQ01004446 JAT35531.1 CVRI01000054 CRL00195.1 GECU01025290 JAS82416.1 ATLV01004846 KE524243 KFB35157.1 GFDF01009921 JAV04163.1 GAMD01002809 JAA98781.1 CH477193 EAT48514.1 ATLV01004847 KFB35159.1 AXCP01002751 ADMH02000081 ETN67872.1 AXCN02000233 APCN01005504 AAAB01008888 EAA44640.4 EGK97065.1 UFQT01000091 SSX19532.1 AXCM01001479 KQ765222 OAD54065.1 GALA01000673 JAA94179.1 GFDL01004757 JAV30288.1 DS231953 EDS28787.1 KQ435821 KOX72291.1 KQ435180 KZC14979.1 LBMM01002304 KMQ95011.1 GL437663 EFN70123.1 QOIP01000014 RLU15047.1 KK107083 EZA60083.1 GL767538 EFZ13487.1 KQ977991 KYM98283.1 GL888344 EGI62397.1 GBRD01011404 GBRD01011403 GBRD01010999 JAG54420.1 GDHC01016332 JAQ02297.1 GBHO01020401 JAG23203.1 GBRD01011400 JAG54424.1 GL449965 EFN81694.1 NNAY01001541 OXU23637.1 ADTU01009725 AAZX01000360 AAZX01011714 AAZX01013565 AAZX01016629 AAZX01025833 KQ976428 KYM87825.1 CH916373 EDV94787.1 CM000070 EAL28550.2 CH479182 EDW34334.1 KQ982169 KYQ59534.1 CH964272 EDW84759.1 GAKP01005141 JAC53811.1 CP012526 ALC46635.1 CH933806 EDW15872.1 ABLF02035568 ABLF02035571 JXJN01016575 GDHF01004677 JAI47637.1 CCAG010014772 CH902617 EDV44340.1 GBXI01015869 JAC98422.1 CH940652 EDW59689.1 GFXV01003522 MBW15327.1 GGMR01007089 MBY19708.1 BT120151 ADB57072.1 GAMC01015415 GAMC01015413 JAB91142.1 OUUW01000008 SPP84616.1 GGMS01014366 MBY83569.1 CM000364 EDX11687.1 CH480821 EDW54916.1 CH954181 EDV47854.1 DQ333317 DQ324736 AE014297 AY060630 CM000160 EDW95737.1 REGN01000043 RNA44944.1 GL732612 EFX71189.1 GDIP01249903 JAI73498.1 GDIP01083969 LRGB01002121 JAM19746.1 KZS09027.1 GDIQ01255989 JAJ95735.1 GDIP01007450 JAM96265.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053240
UP000053268
UP000242457
+ More
UP000245037 UP000027135 UP000092461 UP000069940 UP000249989 UP000183832 UP000030765 UP000079169 UP000008820 UP000075880 UP000069272 UP000000673 UP000075886 UP000075840 UP000075903 UP000075902 UP000007062 UP000075883 UP000075920 UP000075884 UP000076407 UP000075900 UP000075881 UP000075885 UP000075901 UP000002320 UP000005203 UP000076408 UP000053105 UP000076502 UP000036403 UP000000311 UP000279307 UP000053097 UP000078542 UP000007755 UP000008237 UP000215335 UP000005205 UP000002358 UP000078540 UP000001070 UP000001819 UP000008744 UP000075809 UP000095300 UP000007798 UP000092553 UP000009192 UP000092445 UP000007819 UP000092443 UP000078200 UP000092460 UP000092444 UP000007801 UP000091820 UP000008792 UP000095301 UP000268350 UP000000304 UP000001292 UP000008711 UP000000803 UP000192221 UP000002282 UP000276133 UP000000305 UP000076858
UP000245037 UP000027135 UP000092461 UP000069940 UP000249989 UP000183832 UP000030765 UP000079169 UP000008820 UP000075880 UP000069272 UP000000673 UP000075886 UP000075840 UP000075903 UP000075902 UP000007062 UP000075883 UP000075920 UP000075884 UP000076407 UP000075900 UP000075881 UP000075885 UP000075901 UP000002320 UP000005203 UP000076408 UP000053105 UP000076502 UP000036403 UP000000311 UP000279307 UP000053097 UP000078542 UP000007755 UP000008237 UP000215335 UP000005205 UP000002358 UP000078540 UP000001070 UP000001819 UP000008744 UP000075809 UP000095300 UP000007798 UP000092553 UP000009192 UP000092445 UP000007819 UP000092443 UP000078200 UP000092460 UP000092444 UP000007801 UP000091820 UP000008792 UP000095301 UP000268350 UP000000304 UP000001292 UP000008711 UP000000803 UP000192221 UP000002282 UP000276133 UP000000305 UP000076858
Pfam
Interpro
IPR003409
MORN
+ More
IPR029240 MMS19_N
IPR024687 MMS19_C
IPR011989 ARM-like
IPR039920 MET18/MMS19
IPR016024 ARM-type_fold
IPR027417 P-loop_NTPase
IPR006297 EF-4
IPR035647 EFG_III/V
IPR009000 Transl_B-barrel_sf
IPR000795 TF_GTP-bd_dom
IPR000640 EFG_V-like
IPR038363 LepA_C_sf
IPR005225 Small_GTP-bd_dom
IPR031157 G_TR_CS
IPR013842 LepA_CTD
IPR022727 Cuticle_C1
IPR029240 MMS19_N
IPR024687 MMS19_C
IPR011989 ARM-like
IPR039920 MET18/MMS19
IPR016024 ARM-type_fold
IPR027417 P-loop_NTPase
IPR006297 EF-4
IPR035647 EFG_III/V
IPR009000 Transl_B-barrel_sf
IPR000795 TF_GTP-bd_dom
IPR000640 EFG_V-like
IPR038363 LepA_C_sf
IPR005225 Small_GTP-bd_dom
IPR031157 G_TR_CS
IPR013842 LepA_CTD
IPR022727 Cuticle_C1
Gene 3D
ProteinModelPortal
H9IVL8
A0A3S2L464
A0A212F3Q2
A0A194QM99
A0A194PDK6
A0A2A3EUI6
+ More
A0A2P8Y895 A0A067RF44 A0A1B0CKH2 A0A182GVT9 A0A1B6FD29 A0A1B6MI08 A0A1J1IIZ5 A0A1B6I651 A0A084VB13 A0A1L8DCG9 T1DQ33 A0A1S3CYE0 Q17PB7 A0A084VB15 A0A182JDS1 A0A182F717 W5JU47 A0A182QRQ6 A0A182HZ81 A0A182V8T2 A0A182TW99 Q7PH89 A0A336LN76 A0A182M6J6 A0A182VS08 A0A310SK63 A0A182NSN3 A0A182WVV7 A0A182RZE1 A0A182KGR3 A0A182P0C9 A0A182T8H4 T1D558 A0A1Q3FRW4 B0WIW1 A0A088AF51 A0A182Y7F9 A0A0N0BEY7 A0A154PST9 A0A0J7KXG4 E2A8P4 A0A3L8D3G6 A0A026WVR0 E9J0W7 A0A151ICZ1 F4WTT2 A0A0K8SNN5 A0A146L576 A0A0A9XWF4 A0A0K8SMT0 E2BRK2 A0A232EZA8 A0A158N9N9 K7JBS3 A0A195BPQ8 B4JSP7 Q296A8 B4GFE9 A0A151XGK6 A0A1I8NX35 B4NI67 A0A034WE33 A0A0M4EQK6 B4KCM3 A0A1A9ZVL4 J9K3X1 A0A1A9Y8I5 A0A1A9VAD9 A0A1B0BLV0 A0A0K8W980 A0A1B0FQT6 B3M170 A0A0A1WJ59 A0A1A9WFS8 B4M4X8 A0A2H8TMD5 A0A2S2NRK1 D2NUK8 A0A1I8NF45 W8AYE1 A0A3B0KFU7 A0A2S2R0P9 B4QW88 B4I3X3 B3P2A4 Q9VN91 A0A1W4UWU5 B4PV68 A0A3M7TA09 E9HAI9 A0A0P4WW95 A0A0P5WN34 A0A0P5H1Y6 A0A0P6CJD3
A0A2P8Y895 A0A067RF44 A0A1B0CKH2 A0A182GVT9 A0A1B6FD29 A0A1B6MI08 A0A1J1IIZ5 A0A1B6I651 A0A084VB13 A0A1L8DCG9 T1DQ33 A0A1S3CYE0 Q17PB7 A0A084VB15 A0A182JDS1 A0A182F717 W5JU47 A0A182QRQ6 A0A182HZ81 A0A182V8T2 A0A182TW99 Q7PH89 A0A336LN76 A0A182M6J6 A0A182VS08 A0A310SK63 A0A182NSN3 A0A182WVV7 A0A182RZE1 A0A182KGR3 A0A182P0C9 A0A182T8H4 T1D558 A0A1Q3FRW4 B0WIW1 A0A088AF51 A0A182Y7F9 A0A0N0BEY7 A0A154PST9 A0A0J7KXG4 E2A8P4 A0A3L8D3G6 A0A026WVR0 E9J0W7 A0A151ICZ1 F4WTT2 A0A0K8SNN5 A0A146L576 A0A0A9XWF4 A0A0K8SMT0 E2BRK2 A0A232EZA8 A0A158N9N9 K7JBS3 A0A195BPQ8 B4JSP7 Q296A8 B4GFE9 A0A151XGK6 A0A1I8NX35 B4NI67 A0A034WE33 A0A0M4EQK6 B4KCM3 A0A1A9ZVL4 J9K3X1 A0A1A9Y8I5 A0A1A9VAD9 A0A1B0BLV0 A0A0K8W980 A0A1B0FQT6 B3M170 A0A0A1WJ59 A0A1A9WFS8 B4M4X8 A0A2H8TMD5 A0A2S2NRK1 D2NUK8 A0A1I8NF45 W8AYE1 A0A3B0KFU7 A0A2S2R0P9 B4QW88 B4I3X3 B3P2A4 Q9VN91 A0A1W4UWU5 B4PV68 A0A3M7TA09 E9HAI9 A0A0P4WW95 A0A0P5WN34 A0A0P5H1Y6 A0A0P6CJD3
Ontologies
GO
PANTHER
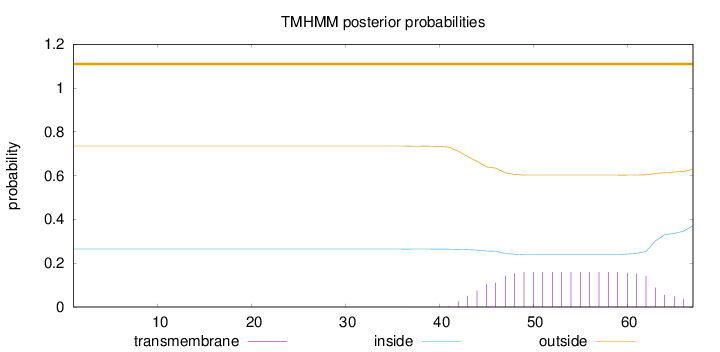
Topology
Subcellular location
Mitochondrion inner membrane
Membrane Co-localizes with ninaC in the rhabdomere membrane. With evidence from 5 publications.
Membrane Co-localizes with ninaC in the rhabdomere membrane. With evidence from 5 publications.
Length:
67
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.10503
Exp number, first 60 AAs:
2.5821
Total prob of N-in:
0.26471
outside
1 - 67
Population Genetic Test Statistics
Pi
246.86557
Theta
121.992525
Tajima's D
3.163516
CLR
0.579061
CSRT
0.986150692465377
Interpretation
Uncertain
