Gene
KWMTBOMO07625
Pre Gene Modal
BGIBMGA001300
Annotation
neuropeptide_receptor_A9_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.981
Sequence
CDS
ATGAATGACCCGGATATAAGCTACAACAGCTCGTTGCTGAGAATGCGCGAGATCACATCGACCCTGGCTCCATTCACGACCATCCGGAATGCGAGTGTGGCGAAGCCTAACAACGCATACGAGTGGCGTTTCATATTACCGCCGTATTTCGTTATATTCCTGCTCTCCATATGCGGGAATTGTCTCGTCATAGCGACTCTAGCGAGCAACAGGCGAATGAGGACAGTCACGAACGTTTATTTACTTAATTTGGCCATTTCGGACTTTTTACTTGGAGTCTTTTGTTTGCCGTTCACGCTCGTCGGACAGATATATAGGAGGTTTCTGTTCGGGGCGGCCCTATGTAAGCTGATACCTTTTCTGCAAGCTGTCTCCGTGTCTGTGGACGTGTGGACATTAGTCGCCATCTCCCTGGAACGATACTTCGCCATCTGTCGCCCGTTGAAGTCCAGGAAATGGCAGACACAATGCCATGCATACAAGATGATAGCCATGGTCTGGATCCTGTCATTGATCCTGAACTCGCCAATCATGCTCGTGTCTACATTGCAGCCAATGAGAGGAAATGGTAAGTTTATTTTTTATTTATTTTTTATTGCTTAG
Protein
MNDPDISYNSSLLRMREITSTLAPFTTIRNASVAKPNNAYEWRFILPPYFVIFLLSICGNCLVIATLASNRRMRTVTNVYLLNLAISDFLLGVFCLPFTLVGQIYRRFLFGAALCKLIPFLQAVSVSVDVWTLVAISLERYFAICRPLKSRKWQTQCHAYKMIAMVWILSLILNSPIMLVSTLQPMRGNGKFIFYLFFIA
Summary
Similarity
Belongs to the G-protein coupled receptor 1 family.
Uniprot
H9IVM0
B3XXM2
A0A0S1YDG2
A0A194QL20
A0A194PCZ3
A0A0L7L806
+ More
A0A182GPU3 A0A182UK86 Q16TK3 Q2TJ53 D2A3E1 M9Z5C3 A0A026WK40 A0A182LIC6 A0A3F2Z1Z4 A0A0M9A2S3 Q7QHX9 B0WGI0 A0A1S4G767 A0A3L8DBR0 A0A0L7QZH6 A0A3F2Z1Z6 Q69DS3 A0A182IDT6 A0A195CBD5 A0A1D2MHN0 A0A154PLJ2 E2BFW0 A0A088A2X9 A0A0P5C943 A0A195BJC4 A0A195DM21 A0A182VB17 A0A195FSZ3 A0A0P5QH23 A0A0P4XNL9 A0A0P5DXR9 A0A0N8ABS5 A0A0N8APA5 A0A0P5I018 A0A0P4ZRY0 A0A0P6FT13 A0A310SF68 A0A0P5X7R1 E9GF26 A0A0P4XKN0 A0A0P5UAP4 A0A0P5BD17 A0A182Y4Z7 A0A0P5U5N1 D2A204 T1IJT3 A0A0P5UHI9 A0A0P5AC41 A0A0P5TQP4 A0A1Y1MGS1 J9HFI9 A0A1S4G641 A0A0P5BCM5 A0A0N8CGQ7 A0A0P5JLP7 A0A2R5L6X1 A0A0P6I6R3 A0A0N8DA49 A0A0P5LSQ3 A0A0P5TJI8 A0A0P4YZU3 A0A336LNY7 A0A0P6C2B0 A0A0P6ITZ5 U3U956 E9GRX7 A0A1B6LSS3 A0A0P5VSU4 A0A0P5WD05 B0XAR6 Q1M2P7 A0A2T7PP38 A0A1I8PI39 B4I6W1 B4NDA6 E0W443 A8JUP8 A8WHF4 B4ND99 B4H7F1 B3NUN0 B5DNQ1 B4M2S9 A0A1W4UQB2 A0A0R1EBR6 A0A0M4EM52 A0A3B0JX14
A0A182GPU3 A0A182UK86 Q16TK3 Q2TJ53 D2A3E1 M9Z5C3 A0A026WK40 A0A182LIC6 A0A3F2Z1Z4 A0A0M9A2S3 Q7QHX9 B0WGI0 A0A1S4G767 A0A3L8DBR0 A0A0L7QZH6 A0A3F2Z1Z6 Q69DS3 A0A182IDT6 A0A195CBD5 A0A1D2MHN0 A0A154PLJ2 E2BFW0 A0A088A2X9 A0A0P5C943 A0A195BJC4 A0A195DM21 A0A182VB17 A0A195FSZ3 A0A0P5QH23 A0A0P4XNL9 A0A0P5DXR9 A0A0N8ABS5 A0A0N8APA5 A0A0P5I018 A0A0P4ZRY0 A0A0P6FT13 A0A310SF68 A0A0P5X7R1 E9GF26 A0A0P4XKN0 A0A0P5UAP4 A0A0P5BD17 A0A182Y4Z7 A0A0P5U5N1 D2A204 T1IJT3 A0A0P5UHI9 A0A0P5AC41 A0A0P5TQP4 A0A1Y1MGS1 J9HFI9 A0A1S4G641 A0A0P5BCM5 A0A0N8CGQ7 A0A0P5JLP7 A0A2R5L6X1 A0A0P6I6R3 A0A0N8DA49 A0A0P5LSQ3 A0A0P5TJI8 A0A0P4YZU3 A0A336LNY7 A0A0P6C2B0 A0A0P6ITZ5 U3U956 E9GRX7 A0A1B6LSS3 A0A0P5VSU4 A0A0P5WD05 B0XAR6 Q1M2P7 A0A2T7PP38 A0A1I8PI39 B4I6W1 B4NDA6 E0W443 A8JUP8 A8WHF4 B4ND99 B4H7F1 B3NUN0 B5DNQ1 B4M2S9 A0A1W4UQB2 A0A0R1EBR6 A0A0M4EM52 A0A3B0JX14
Pubmed
19121390
18725956
26354079
26227816
26483478
17510324
+ More
18362917 19820115 23524001 24508170 20966253 12364791 30249741 14747013 17210077 27289101 20798317 21292972 25244985 28004739 23932938 17994087 20566863 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15632085 17550304
18362917 19820115 23524001 24508170 20966253 12364791 30249741 14747013 17210077 27289101 20798317 21292972 25244985 28004739 23932938 17994087 20566863 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15632085 17550304
EMBL
BABH01001448
BABH01001449
BABH01001450
AB330430
BAG68408.1
KT031007
+ More
ALM88305.1 KQ461198 KPJ06089.1 KQ459606 KPI91097.1 JTDY01002342 KOB71627.1 JXUM01001842 JXUM01001843 JXUM01001844 JXUM01001845 JXUM01001846 KQ560129 KXJ84287.1 CH477646 EAT37847.1 AY865608 AAX56942.1 KQ971338 EFA02835.2 KC161573 AGK29938.1 KK107167 EZA56385.1 KQ435768 KOX75256.1 AAAB01008811 EAA04945.4 DS231926 EDS26978.1 QOIP01000010 RLU17553.1 KQ414680 KOC64029.1 AH013423 AY347947 AY347949 AY347950 AY347951 AAR28375.1 EAU77575.1 APCN01004796 APCN01004797 KQ978009 KYM98174.1 LJIJ01001223 ODM92453.1 KQ434946 KZC12324.1 GL448096 EFN85362.1 GDIP01177896 JAJ45506.1 KQ976455 KYM85204.1 KQ980734 KYN13886.1 KQ981305 KYN43024.1 GDIQ01117254 JAL34472.1 GDIP01238590 JAI84811.1 GDIP01155694 JAJ67708.1 GDIP01162937 JAJ60465.1 GDIQ01256219 JAJ95505.1 GDIQ01220356 JAK31369.1 GDIP01210475 JAJ12927.1 GDIQ01054559 JAN40178.1 KQ760548 OAD59931.1 GDIP01076181 JAM27534.1 GL732541 EFX81934.1 GDIP01242807 JAI80594.1 GDIP01115689 JAL88025.1 GDIP01186229 JAJ37173.1 GDIP01119494 JAL84220.1 EFA02911.1 JH430345 GDIP01117029 JAL86685.1 GDIP01201182 JAJ22220.1 GDIP01124146 JAL79568.1 GEZM01031733 JAV84853.1 CH477351 EJY57597.1 GDIP01186292 JAJ37110.1 GDIP01133391 JAL70323.1 GDIQ01201297 JAK50428.1 GGLE01001130 MBY05256.1 GDIQ01034672 JAN60065.1 GDIP01053855 GDIQ01068002 JAM49860.1 JAN26735.1 GDIQ01165769 JAK85956.1 GDIP01126883 JAL76831.1 GDIP01232042 JAI91359.1 UFQT01000037 SSX18393.1 GDIP01022226 JAM81489.1 GDIQ01005078 JAN89659.1 AB817292 BAO01059.1 GL732561 EFX77608.1 GEBQ01013234 JAT26743.1 GDIP01096096 JAM07619.1 GDIP01087907 JAM15808.1 DS232593 EDS43855.1 AY271617 AAQ01746.1 PZQS01000003 PVD35196.1 CH480823 EDW56059.1 CH964239 EDW82815.2 DS235886 EEB20399.1 AE014298 ABW09450.1 BT031100 ABX00722.1 EDW82808.2 CH479218 EDW33762.1 CH954180 EDV46627.2 CH379065 EDY73152.2 CH940651 EDW65983.2 CM000162 KRK06950.1 CP012528 ALC48721.1 OUUW01000003 SPP77896.1
ALM88305.1 KQ461198 KPJ06089.1 KQ459606 KPI91097.1 JTDY01002342 KOB71627.1 JXUM01001842 JXUM01001843 JXUM01001844 JXUM01001845 JXUM01001846 KQ560129 KXJ84287.1 CH477646 EAT37847.1 AY865608 AAX56942.1 KQ971338 EFA02835.2 KC161573 AGK29938.1 KK107167 EZA56385.1 KQ435768 KOX75256.1 AAAB01008811 EAA04945.4 DS231926 EDS26978.1 QOIP01000010 RLU17553.1 KQ414680 KOC64029.1 AH013423 AY347947 AY347949 AY347950 AY347951 AAR28375.1 EAU77575.1 APCN01004796 APCN01004797 KQ978009 KYM98174.1 LJIJ01001223 ODM92453.1 KQ434946 KZC12324.1 GL448096 EFN85362.1 GDIP01177896 JAJ45506.1 KQ976455 KYM85204.1 KQ980734 KYN13886.1 KQ981305 KYN43024.1 GDIQ01117254 JAL34472.1 GDIP01238590 JAI84811.1 GDIP01155694 JAJ67708.1 GDIP01162937 JAJ60465.1 GDIQ01256219 JAJ95505.1 GDIQ01220356 JAK31369.1 GDIP01210475 JAJ12927.1 GDIQ01054559 JAN40178.1 KQ760548 OAD59931.1 GDIP01076181 JAM27534.1 GL732541 EFX81934.1 GDIP01242807 JAI80594.1 GDIP01115689 JAL88025.1 GDIP01186229 JAJ37173.1 GDIP01119494 JAL84220.1 EFA02911.1 JH430345 GDIP01117029 JAL86685.1 GDIP01201182 JAJ22220.1 GDIP01124146 JAL79568.1 GEZM01031733 JAV84853.1 CH477351 EJY57597.1 GDIP01186292 JAJ37110.1 GDIP01133391 JAL70323.1 GDIQ01201297 JAK50428.1 GGLE01001130 MBY05256.1 GDIQ01034672 JAN60065.1 GDIP01053855 GDIQ01068002 JAM49860.1 JAN26735.1 GDIQ01165769 JAK85956.1 GDIP01126883 JAL76831.1 GDIP01232042 JAI91359.1 UFQT01000037 SSX18393.1 GDIP01022226 JAM81489.1 GDIQ01005078 JAN89659.1 AB817292 BAO01059.1 GL732561 EFX77608.1 GEBQ01013234 JAT26743.1 GDIP01096096 JAM07619.1 GDIP01087907 JAM15808.1 DS232593 EDS43855.1 AY271617 AAQ01746.1 PZQS01000003 PVD35196.1 CH480823 EDW56059.1 CH964239 EDW82815.2 DS235886 EEB20399.1 AE014298 ABW09450.1 BT031100 ABX00722.1 EDW82808.2 CH479218 EDW33762.1 CH954180 EDV46627.2 CH379065 EDY73152.2 CH940651 EDW65983.2 CM000162 KRK06950.1 CP012528 ALC48721.1 OUUW01000003 SPP77896.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000037510
UP000069940
UP000249989
+ More
UP000075902 UP000008820 UP000007266 UP000053097 UP000075882 UP000076407 UP000053105 UP000007062 UP000002320 UP000279307 UP000053825 UP000075840 UP000078542 UP000094527 UP000076502 UP000008237 UP000005203 UP000078540 UP000078492 UP000075903 UP000078541 UP000000305 UP000076408 UP000245119 UP000095300 UP000001292 UP000007798 UP000009046 UP000000803 UP000008744 UP000008711 UP000001819 UP000008792 UP000192221 UP000002282 UP000092553 UP000268350
UP000075902 UP000008820 UP000007266 UP000053097 UP000075882 UP000076407 UP000053105 UP000007062 UP000002320 UP000279307 UP000053825 UP000075840 UP000078542 UP000094527 UP000076502 UP000008237 UP000005203 UP000078540 UP000078492 UP000075903 UP000078541 UP000000305 UP000076408 UP000245119 UP000095300 UP000001292 UP000007798 UP000009046 UP000000803 UP000008744 UP000008711 UP000001819 UP000008792 UP000192221 UP000002282 UP000092553 UP000268350
Pfam
PF00001 7tm_1
Interpro
ProteinModelPortal
H9IVM0
B3XXM2
A0A0S1YDG2
A0A194QL20
A0A194PCZ3
A0A0L7L806
+ More
A0A182GPU3 A0A182UK86 Q16TK3 Q2TJ53 D2A3E1 M9Z5C3 A0A026WK40 A0A182LIC6 A0A3F2Z1Z4 A0A0M9A2S3 Q7QHX9 B0WGI0 A0A1S4G767 A0A3L8DBR0 A0A0L7QZH6 A0A3F2Z1Z6 Q69DS3 A0A182IDT6 A0A195CBD5 A0A1D2MHN0 A0A154PLJ2 E2BFW0 A0A088A2X9 A0A0P5C943 A0A195BJC4 A0A195DM21 A0A182VB17 A0A195FSZ3 A0A0P5QH23 A0A0P4XNL9 A0A0P5DXR9 A0A0N8ABS5 A0A0N8APA5 A0A0P5I018 A0A0P4ZRY0 A0A0P6FT13 A0A310SF68 A0A0P5X7R1 E9GF26 A0A0P4XKN0 A0A0P5UAP4 A0A0P5BD17 A0A182Y4Z7 A0A0P5U5N1 D2A204 T1IJT3 A0A0P5UHI9 A0A0P5AC41 A0A0P5TQP4 A0A1Y1MGS1 J9HFI9 A0A1S4G641 A0A0P5BCM5 A0A0N8CGQ7 A0A0P5JLP7 A0A2R5L6X1 A0A0P6I6R3 A0A0N8DA49 A0A0P5LSQ3 A0A0P5TJI8 A0A0P4YZU3 A0A336LNY7 A0A0P6C2B0 A0A0P6ITZ5 U3U956 E9GRX7 A0A1B6LSS3 A0A0P5VSU4 A0A0P5WD05 B0XAR6 Q1M2P7 A0A2T7PP38 A0A1I8PI39 B4I6W1 B4NDA6 E0W443 A8JUP8 A8WHF4 B4ND99 B4H7F1 B3NUN0 B5DNQ1 B4M2S9 A0A1W4UQB2 A0A0R1EBR6 A0A0M4EM52 A0A3B0JX14
A0A182GPU3 A0A182UK86 Q16TK3 Q2TJ53 D2A3E1 M9Z5C3 A0A026WK40 A0A182LIC6 A0A3F2Z1Z4 A0A0M9A2S3 Q7QHX9 B0WGI0 A0A1S4G767 A0A3L8DBR0 A0A0L7QZH6 A0A3F2Z1Z6 Q69DS3 A0A182IDT6 A0A195CBD5 A0A1D2MHN0 A0A154PLJ2 E2BFW0 A0A088A2X9 A0A0P5C943 A0A195BJC4 A0A195DM21 A0A182VB17 A0A195FSZ3 A0A0P5QH23 A0A0P4XNL9 A0A0P5DXR9 A0A0N8ABS5 A0A0N8APA5 A0A0P5I018 A0A0P4ZRY0 A0A0P6FT13 A0A310SF68 A0A0P5X7R1 E9GF26 A0A0P4XKN0 A0A0P5UAP4 A0A0P5BD17 A0A182Y4Z7 A0A0P5U5N1 D2A204 T1IJT3 A0A0P5UHI9 A0A0P5AC41 A0A0P5TQP4 A0A1Y1MGS1 J9HFI9 A0A1S4G641 A0A0P5BCM5 A0A0N8CGQ7 A0A0P5JLP7 A0A2R5L6X1 A0A0P6I6R3 A0A0N8DA49 A0A0P5LSQ3 A0A0P5TJI8 A0A0P4YZU3 A0A336LNY7 A0A0P6C2B0 A0A0P6ITZ5 U3U956 E9GRX7 A0A1B6LSS3 A0A0P5VSU4 A0A0P5WD05 B0XAR6 Q1M2P7 A0A2T7PP38 A0A1I8PI39 B4I6W1 B4NDA6 E0W443 A8JUP8 A8WHF4 B4ND99 B4H7F1 B3NUN0 B5DNQ1 B4M2S9 A0A1W4UQB2 A0A0R1EBR6 A0A0M4EM52 A0A3B0JX14
PDB
5WS3
E-value=1.80686e-27,
Score=301
Ontologies
KEGG
GO
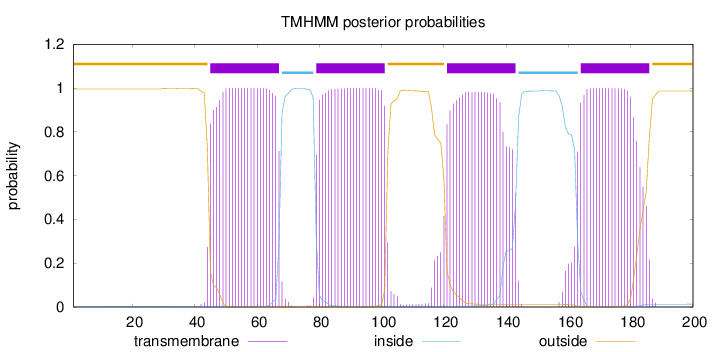
Topology
Length:
200
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
89.9579
Exp number, first 60 AAs:
15.954
Total prob of N-in:
0.00389
POSSIBLE N-term signal
sequence
outside
1 - 44
TMhelix
45 - 67
inside
68 - 78
TMhelix
79 - 101
outside
102 - 120
TMhelix
121 - 143
inside
144 - 163
TMhelix
164 - 186
outside
187 - 200
Population Genetic Test Statistics
Pi
203.546472
Theta
132.551711
Tajima's D
0.397652
CLR
0.192989
CSRT
0.478626068696565
Interpretation
Uncertain
