Gene
KWMTBOMO07623
Pre Gene Modal
BGIBMGA000841
Annotation
PREDICTED:_transmembrane_protease_serine_9-like_[Bombyx_mori]
Full name
Trypsin-3
+ More
Trypsin-2
Trypsin-1
Trypsin-7
Trypsin-2
Trypsin-1
Trypsin-7
Alternative Name
Antryp2
Antryp1
Antryp1
Location in the cell
Cytoplasmic Reliability : 2.972
Sequence
CDS
ATGGGCATTCTTAAGAAAAAAATGGCCAGTGCCGTCAAGGTCGGTACCTTCGATTTGTCCAGACTTGTTTTGAACGAGGAATTAGATGAAGACGAATCATTACTCGTAGATACGGCAGATAGTAGGATTGTGGGTGGACATGAAACTACTATTGAGGACCATCCTTATCAAGTGTCATTCATTGTCAACAACTCGTATTTTTGTGGTGGCATAATCTTAAGTCAAGATTGGATTATGACCGCCGCACATTGCGCTCAAAATGTCGACCCTACAACAGTGGTGCTGCGGGCTGGTAGCACCTTCCGGAAAAATGGCACCATAATACCAATAGCTTTAGTAGCTAGTCATCCAGAATACGATGACCCTGCTTTCGACAAAGACGTTGCTGTCATGAAAACAGTGAAACCCATGGAGTTTGGAGAGACAATGCAGCCGGTAGCACTGCCAGCTCTGGGGGCGAAGTTGGAGCCGAATTCTGAAATCGTAGTCAGTGGTTGGGGGAGGACAGTGATGGGATAA
Protein
MGILKKKMASAVKVGTFDLSRLVLNEELDEDESLLVDTADSRIVGGHETTIEDHPYQVSFIVNNSYFCGGIILSQDWIMTAAHCAQNVDPTTVVLRAGSTFRKNGTIIPIALVASHPEYDDPAFDKDVAVMKTVKPMEFGETMQPVALPALGAKLEPNSEIVVSGWGRTVMG
Summary
Description
Constitutive trypsin that is expressed 2 days after emergence, coinciding with host seeking behavior of the female.
Major function may be to aid in digestion of the blood meal.
Major function may be to aid in digestion of the blood meal.
Catalytic Activity
Preferential cleavage: Arg-|-Xaa, Lys-|-Xaa.
Similarity
Belongs to the peptidase S1 family.
Keywords
Complete proteome
Digestion
Disulfide bond
Hydrolase
Protease
Reference proteome
Secreted
Serine protease
Signal
Zymogen
Feature
propeptide Activation peptide
chain Trypsin-3
chain Trypsin-3
Uniprot
H9IUB1
A0A2H1WLN2
H9IUB0
A0A2A4K471
I3UIJ2
A0A3S2LFV6
+ More
A0A194PDK1 A0A0N1PJX5 A0A212FPE2 A0A0L7KY81 A0A194QL10 A0A194PCY4 A0A194PEH2 A0A0N1IQS6 A0A212EWZ2 H9JW07 A0A2H1X0F7 A0A182GAC9 Q5QBG0 A0A1Y9GLX5 Q16PR8 F4MI42 P35037 A0A1S4GXU9 A0A182V8D5 A0A182TJQ8 A0A182U9U3 A0A1Y9J184 A0A182KHH7 A0A1L9WNS2 Q5QBG2 A0A182VGJ0 A0A2C9GTQ1 P35036 A0A1Y9GKV0 A0A1Y9J0J4 A0A2V5J8A4 A0A1Y9J180 P35035 A0A240PM69 A0A1S4H7M9 A0A0A2KWH5 Q7Q344 A0A182UPH4 A0A182LPG3 A0A340TBF8 C5IB65 A0A084VY05 B0XB10 C5IBB9 A0A194PDG5 A0A182UGL3 A0A182U4C7 C5IBC0 A0A084VY04 C5IBB8 B0W771 A0A182XL71 A0A3D8T2K0 C5IB66 P35041 C5IBF5 W5JDQ9 K7J8G2 A0A182ICF6 A0A084VY03 A0A2V5HHZ2 L0ATN6 A0A182FPP1 C5IB64 C5IB79 A0A2C9GTS0 C5IB68 A0A194RI37 A0A182V8R1 C5IB72 C5IBE8 A0A2A3E9B5 A0A182T724 A0A240PL37 Q29MJ9 J3JW59 C5IB70 A0A182YPE1 A0A1Q3FNA4 A0A2C9GTY9 N6TW63 B0WFK6 A0A1Q3FPS2 C5IBF3 C5IB77 U4TXR1 A0A1Y9GKU7 A0A182WCM2 A0A182TKI7 Q16TM1 A0A023EM39
A0A194PDK1 A0A0N1PJX5 A0A212FPE2 A0A0L7KY81 A0A194QL10 A0A194PCY4 A0A194PEH2 A0A0N1IQS6 A0A212EWZ2 H9JW07 A0A2H1X0F7 A0A182GAC9 Q5QBG0 A0A1Y9GLX5 Q16PR8 F4MI42 P35037 A0A1S4GXU9 A0A182V8D5 A0A182TJQ8 A0A182U9U3 A0A1Y9J184 A0A182KHH7 A0A1L9WNS2 Q5QBG2 A0A182VGJ0 A0A2C9GTQ1 P35036 A0A1Y9GKV0 A0A1Y9J0J4 A0A2V5J8A4 A0A1Y9J180 P35035 A0A240PM69 A0A1S4H7M9 A0A0A2KWH5 Q7Q344 A0A182UPH4 A0A182LPG3 A0A340TBF8 C5IB65 A0A084VY05 B0XB10 C5IBB9 A0A194PDG5 A0A182UGL3 A0A182U4C7 C5IBC0 A0A084VY04 C5IBB8 B0W771 A0A182XL71 A0A3D8T2K0 C5IB66 P35041 C5IBF5 W5JDQ9 K7J8G2 A0A182ICF6 A0A084VY03 A0A2V5HHZ2 L0ATN6 A0A182FPP1 C5IB64 C5IB79 A0A2C9GTS0 C5IB68 A0A194RI37 A0A182V8R1 C5IB72 C5IBE8 A0A2A3E9B5 A0A182T724 A0A240PL37 Q29MJ9 J3JW59 C5IB70 A0A182YPE1 A0A1Q3FNA4 A0A2C9GTY9 N6TW63 B0WFK6 A0A1Q3FPS2 C5IBF3 C5IB77 U4TXR1 A0A1Y9GKU7 A0A182WCM2 A0A182TKI7 Q16TM1 A0A023EM39
EC Number
3.4.21.4
Pubmed
EMBL
BABH01001457
BABH01001458
ODYU01009503
SOQ53993.1
BABH01001460
BABH01001461
+ More
NWSH01000168 PCG78826.1 JN315766 AFK64830.1 RSAL01000017 RVE52910.1 KQ459606 KPI91088.1 LADJ01066212 KPJ21686.1 AGBW02003040 OWR55583.1 JTDY01004566 KOB68004.1 KQ461198 KPJ06079.1 KPI91087.1 KPI91089.1 KPJ21685.1 AGBW02011879 OWR45991.1 BABH01036516 ODYU01012498 SOQ58840.1 JXUM01008849 KQ560270 KXJ83415.1 AY752851 AAV84264.1 APCN01000798 CH477774 EAT36354.1 EZ933290 ADJ57671.1 Z22930 AAAB01008964 KV878981 OJJ97808.1 AY752849 AAV84262.1 Z18890 KZ825476 PYI34337.1 Z18889 AAAB01008966 JQGA01001183 KGO68710.1 EAA13086.4 FJ839225 ACR61236.1 ATLV01018235 ATLV01018236 KE525226 KFB42849.1 DS232607 EDS43980.1 FJ839309 ACR61290.1 KPI91053.1 FJ839310 ACR61291.1 KFB42848.1 FJ839308 FJ839341 ACR61289.1 ACR61322.1 DS231852 EDS37676.1 PVWQ01000001 RDW92774.1 FJ839226 ACR61237.1 FJ839345 ACR61326.1 ADMH02001669 ETN61483.1 KFB42847.1 KZ825103 PYI24068.1 JX915894 AFZ78860.1 FJ839224 ACR61235.1 FJ839239 FJ839240 FJ839241 FJ839242 FJ839244 FJ839245 ACR61250.1 ACR61251.1 ACR61252.1 ACR61253.1 ACR61255.1 ACR61256.1 FJ839228 FJ839231 ACR61239.1 ACR61242.1 KQ460205 KPJ17099.1 FJ839232 FJ839233 FJ839234 FJ839235 FJ839236 FJ839238 ACR61243.1 ACR61244.1 ACR61245.1 ACR61246.1 ACR61247.1 ACR61249.1 FJ839338 ACR61319.1 KZ288316 PBC28310.1 CH379060 EAL33694.2 BT127477 AEE62439.1 FJ839230 ACR61241.1 GFDL01006008 JAV29037.1 APGK01052535 KB741213 ENN72611.1 DS231918 EDS26274.1 GFDL01005572 JAV29473.1 FJ839343 ACR61324.1 FJ839237 ACR61248.1 KB631625 ERL84793.1 CH477645 EAT37856.1 GAPW01003247 JAC10351.1
NWSH01000168 PCG78826.1 JN315766 AFK64830.1 RSAL01000017 RVE52910.1 KQ459606 KPI91088.1 LADJ01066212 KPJ21686.1 AGBW02003040 OWR55583.1 JTDY01004566 KOB68004.1 KQ461198 KPJ06079.1 KPI91087.1 KPI91089.1 KPJ21685.1 AGBW02011879 OWR45991.1 BABH01036516 ODYU01012498 SOQ58840.1 JXUM01008849 KQ560270 KXJ83415.1 AY752851 AAV84264.1 APCN01000798 CH477774 EAT36354.1 EZ933290 ADJ57671.1 Z22930 AAAB01008964 KV878981 OJJ97808.1 AY752849 AAV84262.1 Z18890 KZ825476 PYI34337.1 Z18889 AAAB01008966 JQGA01001183 KGO68710.1 EAA13086.4 FJ839225 ACR61236.1 ATLV01018235 ATLV01018236 KE525226 KFB42849.1 DS232607 EDS43980.1 FJ839309 ACR61290.1 KPI91053.1 FJ839310 ACR61291.1 KFB42848.1 FJ839308 FJ839341 ACR61289.1 ACR61322.1 DS231852 EDS37676.1 PVWQ01000001 RDW92774.1 FJ839226 ACR61237.1 FJ839345 ACR61326.1 ADMH02001669 ETN61483.1 KFB42847.1 KZ825103 PYI24068.1 JX915894 AFZ78860.1 FJ839224 ACR61235.1 FJ839239 FJ839240 FJ839241 FJ839242 FJ839244 FJ839245 ACR61250.1 ACR61251.1 ACR61252.1 ACR61253.1 ACR61255.1 ACR61256.1 FJ839228 FJ839231 ACR61239.1 ACR61242.1 KQ460205 KPJ17099.1 FJ839232 FJ839233 FJ839234 FJ839235 FJ839236 FJ839238 ACR61243.1 ACR61244.1 ACR61245.1 ACR61246.1 ACR61247.1 ACR61249.1 FJ839338 ACR61319.1 KZ288316 PBC28310.1 CH379060 EAL33694.2 BT127477 AEE62439.1 FJ839230 ACR61241.1 GFDL01006008 JAV29037.1 APGK01052535 KB741213 ENN72611.1 DS231918 EDS26274.1 GFDL01005572 JAV29473.1 FJ839343 ACR61324.1 FJ839237 ACR61248.1 KB631625 ERL84793.1 CH477645 EAT37856.1 GAPW01003247 JAC10351.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053240
UP000007151
+ More
UP000037510 UP000069940 UP000249989 UP000075840 UP000008820 UP000007062 UP000075903 UP000075902 UP000076407 UP000075881 UP000184546 UP000075882 UP000248817 UP000075885 UP000030104 UP000030765 UP000002320 UP000256690 UP000000673 UP000002358 UP000249829 UP000069272 UP000242457 UP000075901 UP000001819 UP000076408 UP000019118 UP000030742 UP000075920
UP000037510 UP000069940 UP000249989 UP000075840 UP000008820 UP000007062 UP000075903 UP000075902 UP000076407 UP000075881 UP000184546 UP000075882 UP000248817 UP000075885 UP000030104 UP000030765 UP000002320 UP000256690 UP000000673 UP000002358 UP000249829 UP000069272 UP000242457 UP000075901 UP000001819 UP000076408 UP000019118 UP000030742 UP000075920
Pfam
PF00089 Trypsin
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
H9IUB1
A0A2H1WLN2
H9IUB0
A0A2A4K471
I3UIJ2
A0A3S2LFV6
+ More
A0A194PDK1 A0A0N1PJX5 A0A212FPE2 A0A0L7KY81 A0A194QL10 A0A194PCY4 A0A194PEH2 A0A0N1IQS6 A0A212EWZ2 H9JW07 A0A2H1X0F7 A0A182GAC9 Q5QBG0 A0A1Y9GLX5 Q16PR8 F4MI42 P35037 A0A1S4GXU9 A0A182V8D5 A0A182TJQ8 A0A182U9U3 A0A1Y9J184 A0A182KHH7 A0A1L9WNS2 Q5QBG2 A0A182VGJ0 A0A2C9GTQ1 P35036 A0A1Y9GKV0 A0A1Y9J0J4 A0A2V5J8A4 A0A1Y9J180 P35035 A0A240PM69 A0A1S4H7M9 A0A0A2KWH5 Q7Q344 A0A182UPH4 A0A182LPG3 A0A340TBF8 C5IB65 A0A084VY05 B0XB10 C5IBB9 A0A194PDG5 A0A182UGL3 A0A182U4C7 C5IBC0 A0A084VY04 C5IBB8 B0W771 A0A182XL71 A0A3D8T2K0 C5IB66 P35041 C5IBF5 W5JDQ9 K7J8G2 A0A182ICF6 A0A084VY03 A0A2V5HHZ2 L0ATN6 A0A182FPP1 C5IB64 C5IB79 A0A2C9GTS0 C5IB68 A0A194RI37 A0A182V8R1 C5IB72 C5IBE8 A0A2A3E9B5 A0A182T724 A0A240PL37 Q29MJ9 J3JW59 C5IB70 A0A182YPE1 A0A1Q3FNA4 A0A2C9GTY9 N6TW63 B0WFK6 A0A1Q3FPS2 C5IBF3 C5IB77 U4TXR1 A0A1Y9GKU7 A0A182WCM2 A0A182TKI7 Q16TM1 A0A023EM39
A0A194PDK1 A0A0N1PJX5 A0A212FPE2 A0A0L7KY81 A0A194QL10 A0A194PCY4 A0A194PEH2 A0A0N1IQS6 A0A212EWZ2 H9JW07 A0A2H1X0F7 A0A182GAC9 Q5QBG0 A0A1Y9GLX5 Q16PR8 F4MI42 P35037 A0A1S4GXU9 A0A182V8D5 A0A182TJQ8 A0A182U9U3 A0A1Y9J184 A0A182KHH7 A0A1L9WNS2 Q5QBG2 A0A182VGJ0 A0A2C9GTQ1 P35036 A0A1Y9GKV0 A0A1Y9J0J4 A0A2V5J8A4 A0A1Y9J180 P35035 A0A240PM69 A0A1S4H7M9 A0A0A2KWH5 Q7Q344 A0A182UPH4 A0A182LPG3 A0A340TBF8 C5IB65 A0A084VY05 B0XB10 C5IBB9 A0A194PDG5 A0A182UGL3 A0A182U4C7 C5IBC0 A0A084VY04 C5IBB8 B0W771 A0A182XL71 A0A3D8T2K0 C5IB66 P35041 C5IBF5 W5JDQ9 K7J8G2 A0A182ICF6 A0A084VY03 A0A2V5HHZ2 L0ATN6 A0A182FPP1 C5IB64 C5IB79 A0A2C9GTS0 C5IB68 A0A194RI37 A0A182V8R1 C5IB72 C5IBE8 A0A2A3E9B5 A0A182T724 A0A240PL37 Q29MJ9 J3JW59 C5IB70 A0A182YPE1 A0A1Q3FNA4 A0A2C9GTY9 N6TW63 B0WFK6 A0A1Q3FPS2 C5IBF3 C5IB77 U4TXR1 A0A1Y9GKU7 A0A182WCM2 A0A182TKI7 Q16TM1 A0A023EM39
PDB
5CE1
E-value=4.36192e-15,
Score=193
Ontologies
GO
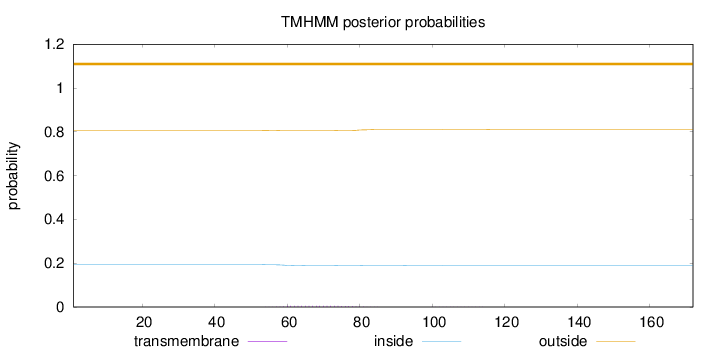
Topology
Subcellular location
Secreted
Length:
172
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11787
Exp number, first 60 AAs:
0.01349
Total prob of N-in:
0.19332
outside
1 - 172
Population Genetic Test Statistics
Pi
230.513823
Theta
186.830722
Tajima's D
0.756583
CLR
0
CSRT
0.590970451477426
Interpretation
Uncertain
