Gene
KWMTBOMO07622
Pre Gene Modal
BGIBMGA000840
Annotation
PREDICTED:_transmembrane_protease_serine_9-like_[Bombyx_mori]
Full name
Trypsin-1
+ More
Trypsin-3
Trypsin-2
Trypsin-7
Trypsin 5G1
Trypsin 3A1
Trypsin-4
Trypsin-3
Trypsin-2
Trypsin-7
Trypsin 5G1
Trypsin 3A1
Trypsin-4
Alternative Name
Antryp1
Antryp2
Antryp2
Location in the cell
Extracellular Reliability : 2.248
Sequence
CDS
ATGGAGTCTTCTGAGCATTTCGTGATTTTCTTTGGATTAACTCTACTATATTCACAACTTGCGTACGGTCTAGATTTCTCCAATTTCCCAACTTTGGAAGAGTTCCTCAAGGAGCCCATGGTAAACGATGACAGAATCGTGGGGGGACAGGAGGCCTTCATCGATGATTACCCCCACCAAGTATCGTTTGTGGTGAACAATACGTATTTTTGCGGCGGCTCGATCATCAGCGAGAACTGGATATTGACTGCAGCCCATTGCTCTCAGAACGTCGACCCCTCGACAGTGGTCTTAAGAGGTGGAAGTTCATGGCGGAAGAATGGTACTATCATTCCCATTGAAAAAGTGATAGCGCATCCAGAGTATGACAACCCTGCGTTCGACAAGGATGTCGCTGTTATGAAGACCAAAGAACCTATACAATTTAGTGACACAATGCAGCCTATAGGGCTACCTTCAATGGATAGAGCAATGAGAGGTGGAACGGAGGTATCGGTCAGCGGGTGGGGACGTACTAAGCAAGGAGCCGCTTCGATTCCCGAAAGACTGATGGACGTGCGTATACCAGTCGTGTCGTATCCGGAGTGTTTCCTGTCGTACTTTTCAGTCCTCACGAGGAACATGTGGTGCGGCGGGAACTTCTTCTTGGGTGGTCAGGGAACTTGCCAGGGCGATTCTGGAGGATCAGCAGTTCAAGACGGTATGGCAGTTGGCATCGTGTCTTTTGGACGTGGTTGTGGCCAGTCTTTCGCTCCAAGTGTGTTCGCCAATATTGCCGCACCTCCCATCAGGAACTTCATCAAAGAACACACAGATCTTTAA
Protein
MESSEHFVIFFGLTLLYSQLAYGLDFSNFPTLEEFLKEPMVNDDRIVGGQEAFIDDYPHQVSFVVNNTYFCGGSIISENWILTAAHCSQNVDPSTVVLRGGSSWRKNGTIIPIEKVIAHPEYDNPAFDKDVAVMKTKEPIQFSDTMQPIGLPSMDRAMRGGTEVSVSGWGRTKQGAASIPERLMDVRIPVVSYPECFLSYFSVLTRNMWCGGNFFLGGQGTCQGDSGGSAVQDGMAVGIVSFGRGCGQSFAPSVFANIAAPPIRNFIKEHTDL
Summary
Description
Major function may be to aid in digestion of the blood meal.
Constitutive trypsin that is expressed 2 days after emergence, coinciding with host seeking behavior of the female.
Constitutive trypsin that is expressed 2 days after emergence, coinciding with host seeking behavior of the female.
Catalytic Activity
Preferential cleavage: Arg-|-Xaa, Lys-|-Xaa.
Similarity
Belongs to the peptidase S1 family.
Keywords
Complete proteome
Digestion
Disulfide bond
Hydrolase
Protease
Reference proteome
Secreted
Serine protease
Signal
Zymogen
Alternative splicing
Feature
propeptide Activation peptide
chain Trypsin-1
splice variant In isoform A.
chain Trypsin-1
splice variant In isoform A.
Uniprot
H9IUB0
A0A2H1WLN2
A0A2A4K471
A0A3S2LFV6
A0A2A4K3K4
A0A2A4K3J2
+ More
H9IUB1 A0A194PDK1 A0A0N1PJX5 A0A212FPE2 A0A0L7KY81 A0A194PEH2 A0A194PCY4 A0A194QL10 A0A0N1IQS6 A0A212EWZ2 A0A2H1X0F7 A0A194PDG5 A0A1Y9J180 P35035 A0A0A1TPQ4 A0A194RI37 A0A2C9GTY9 A0A182U9U3 A0A1Y9GKU7 A0A182TKI7 P35037 A0A1Y9GLX5 A0A182VG56 A0A1S4GXU9 H9JW07 A0A182T760 A0A182V8D5 A0A182TJQ8 A0A1Y9J0J4 A0A1Y9GKV0 P35036 A0A1Y9J184 A0A182VGJ0 A0A2C9GTQ1 L0ATN6 A0A182KHH7 A0A240PL34 A0A0A1T583 P35041 A0A2P0XIZ8 F6JSA6 A0A182V8R1 A0A182ICF6 A0A182T724 A0A182XL71 Q16TM1 A0A182U4C7 A0A0A1TJ15 A0A2C9GTS0 A0A182L0V6 Q5QBF4 A0A182M8Z6 B4KK96 A0A0L7LAG0 Q1M0X9 C8CGF6 C5IBE8 A0A1Y9HES5 Q17086 A0A0A1TC23 A0A182MJH4 A0A1Y9GLA7 A0A1Y9J0S1 O16133 A0A2C9GTS6 P29787 P29786-2 P35038 A0A1V1G554 A0A1Y9IS50 A0A240PM69 A0A240PKC7 F4MI42 A0A084VY03 A0A084VY04 A0A084VY00 A0A2K7P6D6 A0A154PBT0 Q5QBG0 A0A2P0XIE9 C5IBF3 A0A182HBG3 P29786 A0A2M9M4T8 A0A240PL37 D2A2R7 Q16TL9 A0A182G5M7 C5IBF5 K7J8G1 B4LRU7 B4KK93 A0A0B7KIN4
H9IUB1 A0A194PDK1 A0A0N1PJX5 A0A212FPE2 A0A0L7KY81 A0A194PEH2 A0A194PCY4 A0A194QL10 A0A0N1IQS6 A0A212EWZ2 A0A2H1X0F7 A0A194PDG5 A0A1Y9J180 P35035 A0A0A1TPQ4 A0A194RI37 A0A2C9GTY9 A0A182U9U3 A0A1Y9GKU7 A0A182TKI7 P35037 A0A1Y9GLX5 A0A182VG56 A0A1S4GXU9 H9JW07 A0A182T760 A0A182V8D5 A0A182TJQ8 A0A1Y9J0J4 A0A1Y9GKV0 P35036 A0A1Y9J184 A0A182VGJ0 A0A2C9GTQ1 L0ATN6 A0A182KHH7 A0A240PL34 A0A0A1T583 P35041 A0A2P0XIZ8 F6JSA6 A0A182V8R1 A0A182ICF6 A0A182T724 A0A182XL71 Q16TM1 A0A182U4C7 A0A0A1TJ15 A0A2C9GTS0 A0A182L0V6 Q5QBF4 A0A182M8Z6 B4KK96 A0A0L7LAG0 Q1M0X9 C8CGF6 C5IBE8 A0A1Y9HES5 Q17086 A0A0A1TC23 A0A182MJH4 A0A1Y9GLA7 A0A1Y9J0S1 O16133 A0A2C9GTS6 P29787 P29786-2 P35038 A0A1V1G554 A0A1Y9IS50 A0A240PM69 A0A240PKC7 F4MI42 A0A084VY03 A0A084VY04 A0A084VY00 A0A2K7P6D6 A0A154PBT0 Q5QBG0 A0A2P0XIE9 C5IBF3 A0A182HBG3 P29786 A0A2M9M4T8 A0A240PL37 D2A2R7 Q16TL9 A0A182G5M7 C5IBF5 K7J8G1 B4LRU7 B4KK93 A0A0B7KIN4
EC Number
3.4.21.4
Pubmed
EMBL
BABH01001460
BABH01001461
ODYU01009503
SOQ53993.1
NWSH01000168
PCG78826.1
+ More
RSAL01000017 RVE52910.1 PCG78837.1 PCG78825.1 BABH01001457 BABH01001458 KQ459606 KPI91088.1 LADJ01066212 KPJ21686.1 AGBW02003040 OWR55583.1 JTDY01004566 KOB68004.1 KPI91089.1 KPI91087.1 KQ461198 KPJ06079.1 KPJ21685.1 AGBW02011879 OWR45991.1 ODYU01012498 SOQ58840.1 KPI91053.1 Z18889 Z22930 AAAB01008964 CDHN01000005 CEJ92817.1 KQ460205 KPJ17099.1 APCN01000798 BABH01036516 Z18890 JX915894 AFZ78860.1 CDHN01000004 CEJ92321.1 KY661301 AVA17388.1 GU187072 ADQ53625.1 CH477645 EAT37856.1 CDHN01000003 CEJ90625.1 AY752857 AAV84270.1 AXCM01021675 CH933807 EDW12627.1 JTDY01001944 KOB72483.1 AY792954 AAX33734.1 GQ398048 ACV07665.1 FJ839338 ACR61319.1 U52359 AAA97479.1 CEJ92324.1 AXCM01007963 AF012809 AAB66878.1 CH478086 X64363 AF487426 AF508783 X64362 CH477469 FX985313 BAX07326.1 EZ933290 ADJ57671.1 ATLV01018235 KE525226 KFB42847.1 KFB42848.1 ATLV01018234 KFB42844.1 KQ434869 KZC09267.1 AY752851 AAV84264.1 KY661306 AVA17393.1 FJ839343 ACR61324.1 JXUM01032511 KQ560958 KXJ80213.1 NNBP01000013 PJN38624.1 KQ971339 EFA02919.1 EAT37858.1 JXUM01043731 KQ561373 KXJ78808.1 FJ839345 ACR61326.1 CH940649 EDW64699.1 EDW12624.1 CDPU01000050 CEO55277.1
RSAL01000017 RVE52910.1 PCG78837.1 PCG78825.1 BABH01001457 BABH01001458 KQ459606 KPI91088.1 LADJ01066212 KPJ21686.1 AGBW02003040 OWR55583.1 JTDY01004566 KOB68004.1 KPI91089.1 KPI91087.1 KQ461198 KPJ06079.1 KPJ21685.1 AGBW02011879 OWR45991.1 ODYU01012498 SOQ58840.1 KPI91053.1 Z18889 Z22930 AAAB01008964 CDHN01000005 CEJ92817.1 KQ460205 KPJ17099.1 APCN01000798 BABH01036516 Z18890 JX915894 AFZ78860.1 CDHN01000004 CEJ92321.1 KY661301 AVA17388.1 GU187072 ADQ53625.1 CH477645 EAT37856.1 CDHN01000003 CEJ90625.1 AY752857 AAV84270.1 AXCM01021675 CH933807 EDW12627.1 JTDY01001944 KOB72483.1 AY792954 AAX33734.1 GQ398048 ACV07665.1 FJ839338 ACR61319.1 U52359 AAA97479.1 CEJ92324.1 AXCM01007963 AF012809 AAB66878.1 CH478086 X64363 AF487426 AF508783 X64362 CH477469 FX985313 BAX07326.1 EZ933290 ADJ57671.1 ATLV01018235 KE525226 KFB42847.1 KFB42848.1 ATLV01018234 KFB42844.1 KQ434869 KZC09267.1 AY752851 AAV84264.1 KY661306 AVA17393.1 FJ839343 ACR61324.1 JXUM01032511 KQ560958 KXJ80213.1 NNBP01000013 PJN38624.1 KQ971339 EFA02919.1 EAT37858.1 JXUM01043731 KQ561373 KXJ78808.1 FJ839345 ACR61326.1 CH940649 EDW64699.1 EDW12624.1 CDPU01000050 CEO55277.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053240
UP000007151
+ More
UP000037510 UP000076407 UP000007062 UP000039046 UP000075882 UP000075902 UP000075840 UP000075903 UP000075901 UP000075881 UP000075885 UP000008820 UP000075883 UP000009192 UP000075900 UP000030765 UP000076502 UP000069940 UP000249989 UP000231976 UP000007266 UP000002358 UP000008792
UP000037510 UP000076407 UP000007062 UP000039046 UP000075882 UP000075902 UP000075840 UP000075903 UP000075901 UP000075881 UP000075885 UP000008820 UP000075883 UP000009192 UP000075900 UP000030765 UP000076502 UP000069940 UP000249989 UP000231976 UP000007266 UP000002358 UP000008792
Pfam
PF00089 Trypsin
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
H9IUB0
A0A2H1WLN2
A0A2A4K471
A0A3S2LFV6
A0A2A4K3K4
A0A2A4K3J2
+ More
H9IUB1 A0A194PDK1 A0A0N1PJX5 A0A212FPE2 A0A0L7KY81 A0A194PEH2 A0A194PCY4 A0A194QL10 A0A0N1IQS6 A0A212EWZ2 A0A2H1X0F7 A0A194PDG5 A0A1Y9J180 P35035 A0A0A1TPQ4 A0A194RI37 A0A2C9GTY9 A0A182U9U3 A0A1Y9GKU7 A0A182TKI7 P35037 A0A1Y9GLX5 A0A182VG56 A0A1S4GXU9 H9JW07 A0A182T760 A0A182V8D5 A0A182TJQ8 A0A1Y9J0J4 A0A1Y9GKV0 P35036 A0A1Y9J184 A0A182VGJ0 A0A2C9GTQ1 L0ATN6 A0A182KHH7 A0A240PL34 A0A0A1T583 P35041 A0A2P0XIZ8 F6JSA6 A0A182V8R1 A0A182ICF6 A0A182T724 A0A182XL71 Q16TM1 A0A182U4C7 A0A0A1TJ15 A0A2C9GTS0 A0A182L0V6 Q5QBF4 A0A182M8Z6 B4KK96 A0A0L7LAG0 Q1M0X9 C8CGF6 C5IBE8 A0A1Y9HES5 Q17086 A0A0A1TC23 A0A182MJH4 A0A1Y9GLA7 A0A1Y9J0S1 O16133 A0A2C9GTS6 P29787 P29786-2 P35038 A0A1V1G554 A0A1Y9IS50 A0A240PM69 A0A240PKC7 F4MI42 A0A084VY03 A0A084VY04 A0A084VY00 A0A2K7P6D6 A0A154PBT0 Q5QBG0 A0A2P0XIE9 C5IBF3 A0A182HBG3 P29786 A0A2M9M4T8 A0A240PL37 D2A2R7 Q16TL9 A0A182G5M7 C5IBF5 K7J8G1 B4LRU7 B4KK93 A0A0B7KIN4
H9IUB1 A0A194PDK1 A0A0N1PJX5 A0A212FPE2 A0A0L7KY81 A0A194PEH2 A0A194PCY4 A0A194QL10 A0A0N1IQS6 A0A212EWZ2 A0A2H1X0F7 A0A194PDG5 A0A1Y9J180 P35035 A0A0A1TPQ4 A0A194RI37 A0A2C9GTY9 A0A182U9U3 A0A1Y9GKU7 A0A182TKI7 P35037 A0A1Y9GLX5 A0A182VG56 A0A1S4GXU9 H9JW07 A0A182T760 A0A182V8D5 A0A182TJQ8 A0A1Y9J0J4 A0A1Y9GKV0 P35036 A0A1Y9J184 A0A182VGJ0 A0A2C9GTQ1 L0ATN6 A0A182KHH7 A0A240PL34 A0A0A1T583 P35041 A0A2P0XIZ8 F6JSA6 A0A182V8R1 A0A182ICF6 A0A182T724 A0A182XL71 Q16TM1 A0A182U4C7 A0A0A1TJ15 A0A2C9GTS0 A0A182L0V6 Q5QBF4 A0A182M8Z6 B4KK96 A0A0L7LAG0 Q1M0X9 C8CGF6 C5IBE8 A0A1Y9HES5 Q17086 A0A0A1TC23 A0A182MJH4 A0A1Y9GLA7 A0A1Y9J0S1 O16133 A0A2C9GTS6 P29787 P29786-2 P35038 A0A1V1G554 A0A1Y9IS50 A0A240PM69 A0A240PKC7 F4MI42 A0A084VY03 A0A084VY04 A0A084VY00 A0A2K7P6D6 A0A154PBT0 Q5QBG0 A0A2P0XIE9 C5IBF3 A0A182HBG3 P29786 A0A2M9M4T8 A0A240PL37 D2A2R7 Q16TL9 A0A182G5M7 C5IBF5 K7J8G1 B4LRU7 B4KK93 A0A0B7KIN4
PDB
1UTM
E-value=1.80857e-27,
Score=303
Ontologies
GO
Topology
Subcellular location
Secreted
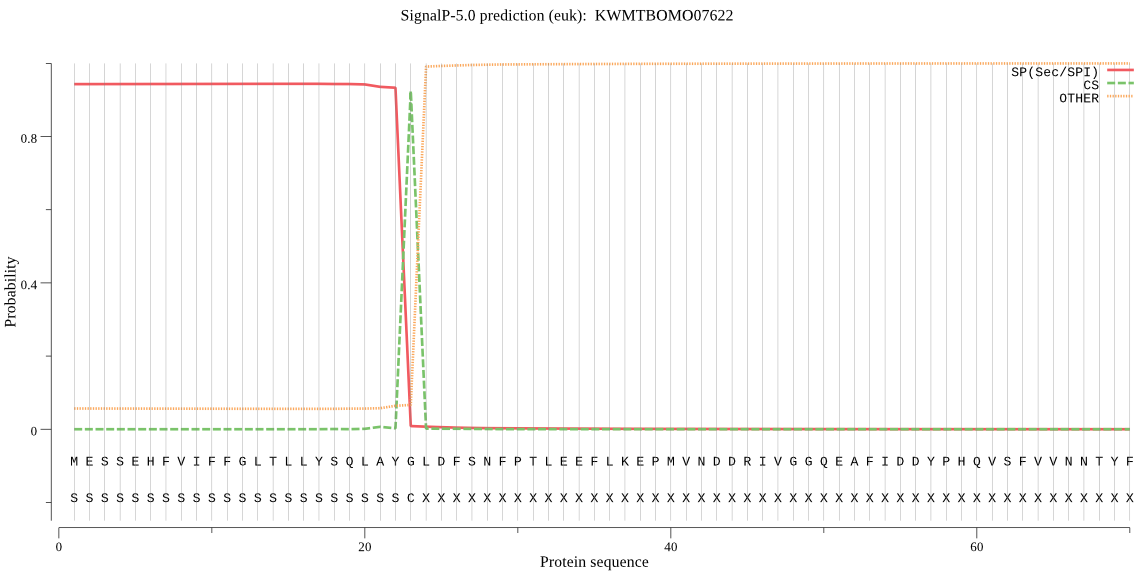
SignalP
Position: 1 - 23,
Likelihood: 0.943490
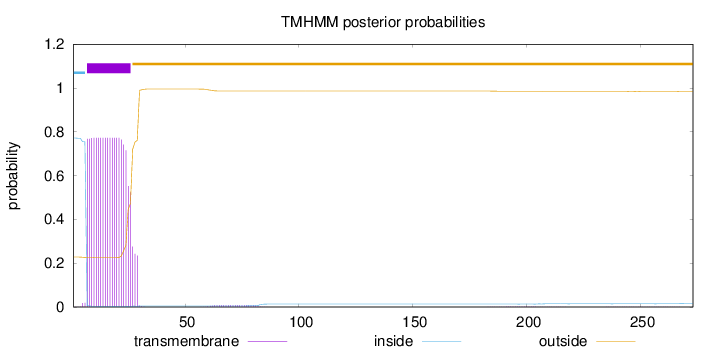
Length:
273
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
15.9279
Exp number, first 60 AAs:
15.68355
Total prob of N-in:
0.77176
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 26
outside
27 - 273
Population Genetic Test Statistics
Pi
189.503383
Theta
160.047429
Tajima's D
1.265117
CLR
0
CSRT
0.729713514324284
Interpretation
Uncertain
