Gene
KWMTBOMO07619 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001302
Annotation
vacuolar_H+_ATP_synthase_16_kDa_proteolipid_subunit_[Bombyx_mori]
Full name
V-type proton ATPase proteolipid subunit
+ More
V-type proton ATPase 16 kDa proteolipid subunit
V-type proton ATPase 16 kDa proteolipid subunit
Alternative Name
Ductin
Vacuolar H+ ATPase subunit 16-1
Vacuolar proton pump 16 kDa proteolipid subunit
V-ATPase subunit C
Vacuolar H+ ATPase subunit 16-1
Vacuolar proton pump 16 kDa proteolipid subunit
V-ATPase subunit C
Location in the cell
PlasmaMembrane Reliability : 3.285
Sequence
CDS
ATGGCTGAAAATAATCCAATCTACGGACCCTTCTTTGGAGTTATGGGGGCGGCGTCTGCTATCATCTTCAGCGCCTTGGGAGCTGCCTATGGAACTGCCAAGTCAGGAACTGGTATTGCCGCCATGTCGGTGATGAGGCCTGAGCTGATCATGAAGTCGATCATTCCTGTCGTCATGGCGGGTATTATTGCCATCTACGGTCTGGTCGTGGCTGTCCTGATTGCTGGTGCCCTCCAGGAGCCAGCCAACTACCCCCTTTACAAAGGGTTCATCCACTTGGGTGCTGGTTTGGCTGTAGGATTCTCTGGTCTGGCTGCCGGTTTCGCCATAGGCATCGTGGGAGATGCAGGCGTGCGTGGTACTGCTCAGCAGCCTAGGAGAGAACATTTCTTTTGCGATTTCTATCGACATATCAGTTTTATTTGCGCGTCACATCCTCCGCAACATCCGAAGATATCAATTTAG
Protein
MAENNPIYGPFFGVMGAASAIIFSALGAAYGTAKSGTGIAAMSVMRPELIMKSIIPVVMAGIIAIYGLVVAVLIAGALQEPANYPLYKGFIHLGAGLAVGFSGLAAGFAIGIVGDAGVRGTAQQPRREHFFCDFYRHISFICASHPPQHPKISI
Summary
Description
Proton-conducting pore forming subunit of the membrane integral V0 complex of vacuolar ATPase. V-ATPase is responsible for acidifying a variety of intracellular compartments in eukaryotic cells.
Subunit
V-ATPase is a heteromultimeric enzyme composed of a peripheral catalytic V1 complex (main components: subunits A, B, C, D, E, and F) attached to an integral membrane V0 proton pore complex (main component: the proteolipid protein; which is present as a hexamer that forms the proton-conducting pore).
Similarity
Belongs to the V-ATPase proteolipid subunit family.
Keywords
Complete proteome
Hydrogen ion transport
Ion transport
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Transport
Vacuole
Feature
chain V-type proton ATPase 16 kDa proteolipid subunit
Uniprot
Q1HQ99
H9IVM2
A0A0P6ILY8
C4WUM0
A0A2H8TS11
D3TLH1
+ More
A0A1B0B1W5 A0A1B0AG93 B4MNV1 A0A2S2P2C6 A0A2M3ZZD6 A0A2M3ZZN9 A0A212F2W5 C4N183 A0A1L8EC69 B4HPV5 B4QD96 A0A1W4VEK7 B3NB54 Q6XI23 P23380 A0A0C9R1H6 I3SJ61 A0A1A9WR00 T1PLX2 X2JC18 A0A3S2M6Z7 A0A2M4A0G7 B3MJ49 W8BRX6 A0A0L0BTS7 T1EAH2 A0A1L8EBZ1 W5JPD7 A0A182J6H0 A0A0K8W669 A0A2M3ZFE7 A0A2M4CJ79 B4LIM5 A0A182P7K6 A0A1Q3FGG5 B0WV62 A0A3B0JL22 A0A1L8DEL5 A8CW60 A0A084WUD4 A0A0A1WRA4 Q28WX5 B4H648 K7J0V3 A0A182YAC2 A0A1S4HCY4 A0A182MKJ3 A0A182QUX3 A0A182WE75 A0A182WXZ1 A0A182FKH1 A0A182RLU4 A0A182I611 A0A182KV05 A0A0M3QVN1 A0A0V0G4Y9 A0A023EFX1 O16110 A0A182NPA9 B4KUH4 A0A232F3Y2 T1DFC6 B4JVQ7 A0A2A4K4J7 A0A2H1VDA2 P55277 A0A1J1IVZ0 A0A2P8ZLX1 R4WNW2 A0A2A3EM58 V9IJP6 Q7YU97 A0A1Y1MP75 P31403 A0A0F7R686 A0A1B1UZP2 A0A1B6MT49 A0A161MMG9 A6YPQ9 A0A2R7X2K5 U5EUW8 A0A164R4R3 I4DM45 A0A194QRB6 A0A0P5I7A0 A0A0P5I961 A0A0N7Z9I6 R4FMV1 A0A069DP10 I4DIU8 Q5QBF6 A0A0P6B0I0 A0A146MBB5 D6X4W4
A0A1B0B1W5 A0A1B0AG93 B4MNV1 A0A2S2P2C6 A0A2M3ZZD6 A0A2M3ZZN9 A0A212F2W5 C4N183 A0A1L8EC69 B4HPV5 B4QD96 A0A1W4VEK7 B3NB54 Q6XI23 P23380 A0A0C9R1H6 I3SJ61 A0A1A9WR00 T1PLX2 X2JC18 A0A3S2M6Z7 A0A2M4A0G7 B3MJ49 W8BRX6 A0A0L0BTS7 T1EAH2 A0A1L8EBZ1 W5JPD7 A0A182J6H0 A0A0K8W669 A0A2M3ZFE7 A0A2M4CJ79 B4LIM5 A0A182P7K6 A0A1Q3FGG5 B0WV62 A0A3B0JL22 A0A1L8DEL5 A8CW60 A0A084WUD4 A0A0A1WRA4 Q28WX5 B4H648 K7J0V3 A0A182YAC2 A0A1S4HCY4 A0A182MKJ3 A0A182QUX3 A0A182WE75 A0A182WXZ1 A0A182FKH1 A0A182RLU4 A0A182I611 A0A182KV05 A0A0M3QVN1 A0A0V0G4Y9 A0A023EFX1 O16110 A0A182NPA9 B4KUH4 A0A232F3Y2 T1DFC6 B4JVQ7 A0A2A4K4J7 A0A2H1VDA2 P55277 A0A1J1IVZ0 A0A2P8ZLX1 R4WNW2 A0A2A3EM58 V9IJP6 Q7YU97 A0A1Y1MP75 P31403 A0A0F7R686 A0A1B1UZP2 A0A1B6MT49 A0A161MMG9 A6YPQ9 A0A2R7X2K5 U5EUW8 A0A164R4R3 I4DM45 A0A194QRB6 A0A0P5I7A0 A0A0P5I961 A0A0N7Z9I6 R4FMV1 A0A069DP10 I4DIU8 Q5QBF6 A0A0P6B0I0 A0A146MBB5 D6X4W4
Pubmed
19121390
20353571
17994087
22118469
19576987
22936249
+ More
18057021 14525923 17550304 2147478 7983150 10731132 12537572 25315136 24528536 24495485 26108605 20920257 23761445 18194529 24438588 25830018 15632085 20075255 25244985 12364791 20966253 24945155 9397516 17510324 28648823 24330624 8353524 29403074 23691247 28004739 1283142 25937057 18207082 22651552 26354079 27129103 26334808 26823975 18362917 19820115
18057021 14525923 17550304 2147478 7983150 10731132 12537572 25315136 24528536 24495485 26108605 20920257 23761445 18194529 24438588 25830018 15632085 20075255 25244985 12364791 20966253 24945155 9397516 17510324 28648823 24330624 8353524 29403074 23691247 28004739 1283142 25937057 18207082 22651552 26354079 27129103 26334808 26823975 18362917 19820115
EMBL
DQ443153
EU082222
ABF51242.1
ABU62758.1
BABH01001468
GDIQ01002357
+ More
JAN92380.1 ABLF02023936 AK341125 BAH71590.1 GFXV01004577 MBW16382.1 EZ422273 ADD18549.1 JXJN01007356 CH963848 EDW73790.1 GGMR01010915 MBY23534.1 GGFK01000603 MBW33924.1 GGFK01000599 MBW33920.1 AGBW02010668 OWR48076.1 EZ048856 ACN69148.1 GFDG01002676 JAV16123.1 CH480816 EDW46624.1 CM000362 CM002911 EDX05883.1 KMY91746.1 CH954177 EDV59819.1 AY232009 CM000157 AAR10032.1 EDW89055.1 X55979 X77936 AE013599 BT012440 GBYB01006717 JAG76484.1 BT140508 AFK40303.1 KA649782 KA649787 AFP64411.1 KJ135006 AHN55086.1 RSAL01000017 RVE52907.1 GGFK01000928 MBW34249.1 CH902619 EDV38143.1 KPU77290.1 GAMC01010544 JAB96011.1 JRES01001352 KNC23406.1 GAMD01001038 JAB00553.1 GFDG01002677 JAV16122.1 ADMH02000484 ETN66257.1 AXCP01005913 GDHF01005752 JAI46562.1 GGFM01006454 MBW27205.1 GGFL01001209 MBW65387.1 CH940648 EDW60395.1 GFDL01008397 JAV26648.1 DS232119 EDS35401.1 OUUW01000001 SPP73341.1 GFDF01009198 JAV04886.1 AJWK01011218 EU124586 ABV60304.1 ATLV01027079 KE525423 KFB53828.1 GBXI01012950 JAD01342.1 CM000071 EAL26541.1 CH479213 EDW33272.1 AAAB01008848 AXCM01001138 AXCN02001819 APCN01003544 APCN01003545 CP012524 ALC42683.1 GECL01003682 JAP02442.1 GAPW01005356 GEHC01001278 JAC08242.1 JAV46367.1 AF008924 CH477190 CH933808 EDW10761.1 NNAY01001086 OXU25188.1 GALA01000672 JAA94180.1 CH916375 EDV98045.1 NWSH01000168 PCG78834.1 ODYU01001899 SOQ38756.1 L16884 CVRI01000063 CRL04443.1 PYGN01000019 PSN57498.1 AK417356 BAN20571.1 KZ288219 PBC32249.1 JR049151 AEY60877.1 AY343324 AAQ21381.1 GEZM01025401 JAV87473.1 X65051 LC026147 BAR72392.1 KU550965 GDQN01003206 ANW09711.1 JAT87848.1 GEBQ01000912 JAT39065.1 GEMB01004294 JAR98978.1 EF639070 ABR27955.1 KK856546 PTY25996.1 GANO01002140 JAB57731.1 LRGB01002230 KZS08322.1 AK402363 BAM18985.1 KQ461198 KPJ06076.1 GDIQ01216823 JAK34902.1 GDIQ01216822 JAK34903.1 GDKW01000443 JAI56152.1 ACPB03011456 GAHY01000823 JAA76687.1 GBGD01003304 JAC85585.1 AK401216 KQ459606 BAM17838.1 KPI91084.1 AY752855 UFQS01000292 UFQT01000292 AAV84268.1 SSX02511.1 SSX22885.1 GDIP01022184 JAM81531.1 GDHC01001558 JAQ17071.1 KQ971381 EEZ97227.1
JAN92380.1 ABLF02023936 AK341125 BAH71590.1 GFXV01004577 MBW16382.1 EZ422273 ADD18549.1 JXJN01007356 CH963848 EDW73790.1 GGMR01010915 MBY23534.1 GGFK01000603 MBW33924.1 GGFK01000599 MBW33920.1 AGBW02010668 OWR48076.1 EZ048856 ACN69148.1 GFDG01002676 JAV16123.1 CH480816 EDW46624.1 CM000362 CM002911 EDX05883.1 KMY91746.1 CH954177 EDV59819.1 AY232009 CM000157 AAR10032.1 EDW89055.1 X55979 X77936 AE013599 BT012440 GBYB01006717 JAG76484.1 BT140508 AFK40303.1 KA649782 KA649787 AFP64411.1 KJ135006 AHN55086.1 RSAL01000017 RVE52907.1 GGFK01000928 MBW34249.1 CH902619 EDV38143.1 KPU77290.1 GAMC01010544 JAB96011.1 JRES01001352 KNC23406.1 GAMD01001038 JAB00553.1 GFDG01002677 JAV16122.1 ADMH02000484 ETN66257.1 AXCP01005913 GDHF01005752 JAI46562.1 GGFM01006454 MBW27205.1 GGFL01001209 MBW65387.1 CH940648 EDW60395.1 GFDL01008397 JAV26648.1 DS232119 EDS35401.1 OUUW01000001 SPP73341.1 GFDF01009198 JAV04886.1 AJWK01011218 EU124586 ABV60304.1 ATLV01027079 KE525423 KFB53828.1 GBXI01012950 JAD01342.1 CM000071 EAL26541.1 CH479213 EDW33272.1 AAAB01008848 AXCM01001138 AXCN02001819 APCN01003544 APCN01003545 CP012524 ALC42683.1 GECL01003682 JAP02442.1 GAPW01005356 GEHC01001278 JAC08242.1 JAV46367.1 AF008924 CH477190 CH933808 EDW10761.1 NNAY01001086 OXU25188.1 GALA01000672 JAA94180.1 CH916375 EDV98045.1 NWSH01000168 PCG78834.1 ODYU01001899 SOQ38756.1 L16884 CVRI01000063 CRL04443.1 PYGN01000019 PSN57498.1 AK417356 BAN20571.1 KZ288219 PBC32249.1 JR049151 AEY60877.1 AY343324 AAQ21381.1 GEZM01025401 JAV87473.1 X65051 LC026147 BAR72392.1 KU550965 GDQN01003206 ANW09711.1 JAT87848.1 GEBQ01000912 JAT39065.1 GEMB01004294 JAR98978.1 EF639070 ABR27955.1 KK856546 PTY25996.1 GANO01002140 JAB57731.1 LRGB01002230 KZS08322.1 AK402363 BAM18985.1 KQ461198 KPJ06076.1 GDIQ01216823 JAK34902.1 GDIQ01216822 JAK34903.1 GDKW01000443 JAI56152.1 ACPB03011456 GAHY01000823 JAA76687.1 GBGD01003304 JAC85585.1 AK401216 KQ459606 BAM17838.1 KPI91084.1 AY752855 UFQS01000292 UFQT01000292 AAV84268.1 SSX02511.1 SSX22885.1 GDIP01022184 JAM81531.1 GDHC01001558 JAQ17071.1 KQ971381 EEZ97227.1
Proteomes
UP000005204
UP000007819
UP000092460
UP000092445
UP000007798
UP000007151
+ More
UP000095300 UP000001292 UP000000304 UP000192221 UP000008711 UP000002282 UP000000803 UP000091820 UP000095301 UP000283053 UP000007801 UP000037069 UP000000673 UP000075880 UP000008792 UP000075885 UP000002320 UP000268350 UP000092461 UP000030765 UP000001819 UP000008744 UP000002358 UP000076408 UP000075883 UP000075886 UP000075920 UP000076407 UP000069272 UP000075900 UP000075840 UP000075882 UP000092553 UP000008820 UP000075884 UP000009192 UP000215335 UP000001070 UP000218220 UP000183832 UP000245037 UP000242457 UP000005203 UP000076858 UP000053240 UP000015103 UP000053268 UP000007266
UP000095300 UP000001292 UP000000304 UP000192221 UP000008711 UP000002282 UP000000803 UP000091820 UP000095301 UP000283053 UP000007801 UP000037069 UP000000673 UP000075880 UP000008792 UP000075885 UP000002320 UP000268350 UP000092461 UP000030765 UP000001819 UP000008744 UP000002358 UP000076408 UP000075883 UP000075886 UP000075920 UP000076407 UP000069272 UP000075900 UP000075840 UP000075882 UP000092553 UP000008820 UP000075884 UP000009192 UP000215335 UP000001070 UP000218220 UP000183832 UP000245037 UP000242457 UP000005203 UP000076858 UP000053240 UP000015103 UP000053268 UP000007266
Pfam
PF00137 ATP-synt_C
Interpro
SUPFAM
SSF81333
SSF81333
ProteinModelPortal
Q1HQ99
H9IVM2
A0A0P6ILY8
C4WUM0
A0A2H8TS11
D3TLH1
+ More
A0A1B0B1W5 A0A1B0AG93 B4MNV1 A0A2S2P2C6 A0A2M3ZZD6 A0A2M3ZZN9 A0A212F2W5 C4N183 A0A1L8EC69 B4HPV5 B4QD96 A0A1W4VEK7 B3NB54 Q6XI23 P23380 A0A0C9R1H6 I3SJ61 A0A1A9WR00 T1PLX2 X2JC18 A0A3S2M6Z7 A0A2M4A0G7 B3MJ49 W8BRX6 A0A0L0BTS7 T1EAH2 A0A1L8EBZ1 W5JPD7 A0A182J6H0 A0A0K8W669 A0A2M3ZFE7 A0A2M4CJ79 B4LIM5 A0A182P7K6 A0A1Q3FGG5 B0WV62 A0A3B0JL22 A0A1L8DEL5 A8CW60 A0A084WUD4 A0A0A1WRA4 Q28WX5 B4H648 K7J0V3 A0A182YAC2 A0A1S4HCY4 A0A182MKJ3 A0A182QUX3 A0A182WE75 A0A182WXZ1 A0A182FKH1 A0A182RLU4 A0A182I611 A0A182KV05 A0A0M3QVN1 A0A0V0G4Y9 A0A023EFX1 O16110 A0A182NPA9 B4KUH4 A0A232F3Y2 T1DFC6 B4JVQ7 A0A2A4K4J7 A0A2H1VDA2 P55277 A0A1J1IVZ0 A0A2P8ZLX1 R4WNW2 A0A2A3EM58 V9IJP6 Q7YU97 A0A1Y1MP75 P31403 A0A0F7R686 A0A1B1UZP2 A0A1B6MT49 A0A161MMG9 A6YPQ9 A0A2R7X2K5 U5EUW8 A0A164R4R3 I4DM45 A0A194QRB6 A0A0P5I7A0 A0A0P5I961 A0A0N7Z9I6 R4FMV1 A0A069DP10 I4DIU8 Q5QBF6 A0A0P6B0I0 A0A146MBB5 D6X4W4
A0A1B0B1W5 A0A1B0AG93 B4MNV1 A0A2S2P2C6 A0A2M3ZZD6 A0A2M3ZZN9 A0A212F2W5 C4N183 A0A1L8EC69 B4HPV5 B4QD96 A0A1W4VEK7 B3NB54 Q6XI23 P23380 A0A0C9R1H6 I3SJ61 A0A1A9WR00 T1PLX2 X2JC18 A0A3S2M6Z7 A0A2M4A0G7 B3MJ49 W8BRX6 A0A0L0BTS7 T1EAH2 A0A1L8EBZ1 W5JPD7 A0A182J6H0 A0A0K8W669 A0A2M3ZFE7 A0A2M4CJ79 B4LIM5 A0A182P7K6 A0A1Q3FGG5 B0WV62 A0A3B0JL22 A0A1L8DEL5 A8CW60 A0A084WUD4 A0A0A1WRA4 Q28WX5 B4H648 K7J0V3 A0A182YAC2 A0A1S4HCY4 A0A182MKJ3 A0A182QUX3 A0A182WE75 A0A182WXZ1 A0A182FKH1 A0A182RLU4 A0A182I611 A0A182KV05 A0A0M3QVN1 A0A0V0G4Y9 A0A023EFX1 O16110 A0A182NPA9 B4KUH4 A0A232F3Y2 T1DFC6 B4JVQ7 A0A2A4K4J7 A0A2H1VDA2 P55277 A0A1J1IVZ0 A0A2P8ZLX1 R4WNW2 A0A2A3EM58 V9IJP6 Q7YU97 A0A1Y1MP75 P31403 A0A0F7R686 A0A1B1UZP2 A0A1B6MT49 A0A161MMG9 A6YPQ9 A0A2R7X2K5 U5EUW8 A0A164R4R3 I4DM45 A0A194QRB6 A0A0P5I7A0 A0A0P5I961 A0A0N7Z9I6 R4FMV1 A0A069DP10 I4DIU8 Q5QBF6 A0A0P6B0I0 A0A146MBB5 D6X4W4
PDB
6O7X
E-value=7.64276e-37,
Score=380
Ontologies
PATHWAY
GO
Topology
Subcellular location
Vacuole membrane
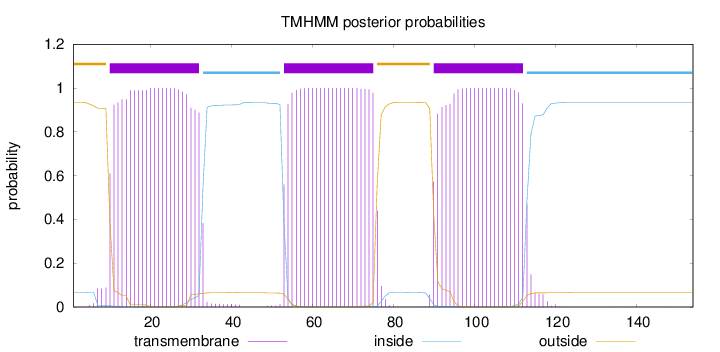
Length:
154
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
68.75188
Exp number, first 60 AAs:
30.22545
Total prob of N-in:
0.06610
POSSIBLE N-term signal
sequence
outside
1 - 9
TMhelix
10 - 32
inside
33 - 52
TMhelix
53 - 75
outside
76 - 89
TMhelix
90 - 112
inside
113 - 154
Population Genetic Test Statistics
Pi
187.176465
Theta
171.733007
Tajima's D
-0.911383
CLR
0.088495
CSRT
0.151442427878606
Interpretation
Uncertain
