Pre Gene Modal
BGIBMGA001303
Annotation
PREDICTED:_tumor_suppressor_candidate_3_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.506
Sequence
CDS
ATGAATTTCAAAGTATTATTGTTACTAACAATAACTATAACATACTATGAAGGAGCTGCGCAACCTCGAGCTAAAGGGATTGAGGAGAAAGTGCAGCAGTTGACTGATATTACAGCAAAGAAGTCTGTAATTCCATTAAACATAAATAAGTTTAAGGAATATGTGAAGTCTCCTCCAAGGGATTATTCATTTGTAGTGATGTTCACTGCAATGGCACCTGCAAGAAGGTGTGCTATCTGTCAGCATGTTAACGATGAATATTTGTTGGTGGCCAACTCATTTAGATTCTCAGCTGCTTACAATAATAAATTATTCTTTGGTATTGTTGATTTTGATGAAGGCTCAGATATCTTCCAAATGTTACGGCTTAATACTGCACCTGTGATAATGCACTTCCCAGCTAAAGGCAAACCTAAGCCTGCTGACACAATGGACTTTGAGAGAGCTGGCATCCATGCTGAGGCCATCGCTAAGTGGATACAGGACAGAACTGATGTACAGATTCGAGTCTTCCGATCACCAAATTACTCAGCGGCTGTAGCTTTTTCAACTCTCTTCATCATATTGGCTGGTTTTCTTTATATCCGTAGAAACAACCTCGAGTTCCTCTACAATAAACAGTTATGGGCAGTTTGTGCTGTGTTCTTCTGCTTTGCAATGGTGTCTGGGCAGATGTGGAATCAAATTAGAGGCCCACCATTTTTCCATAGAACAAAAACTGGCCCGGTCTATATAAACGGGGGATCACACGGACAGTTTGTATTGGAGAGTTACATTGTTGCTATACTGAATGGGGCTGTGGTTGTGGGTATGATCCTAATGATTGAAGCAGCAGGTGGAGTTAAAAACCATGACCCAACACGAACCCTGGCCGAAGGGAAGCGTCGAAGGTTCCAGTCAGTTGTTGGTCTAATATTAGTCTGCGTGTTCTTCTCACTGCTTTTATCAGTATTCAGAGCAAAGACGCAAGGATATCCATATAGTTTCTTGTTCAAGTGA
Protein
MNFKVLLLLTITITYYEGAAQPRAKGIEEKVQQLTDITAKKSVIPLNINKFKEYVKSPPRDYSFVVMFTAMAPARRCAICQHVNDEYLLVANSFRFSAAYNNKLFFGIVDFDEGSDIFQMLRLNTAPVIMHFPAKGKPKPADTMDFERAGIHAEAIAKWIQDRTDVQIRVFRSPNYSAAVAFSTLFIILAGFLYIRRNNLEFLYNKQLWAVCAVFFCFAMVSGQMWNQIRGPPFFHRTKTGPVYINGGSHGQFVLESYIVAILNGAVVVGMILMIEAAGGVKNHDPTRTLAEGKRRRFQSVVGLILVCVFFSLLLSVFRAKTQGYPYSFLFK
Summary
Similarity
Belongs to the insulin family.
Uniprot
H9IVM3
A0A3S2NJZ2
A0A2H1VD56
A0A2A4K4Y6
S4PKA2
I4DK11
+ More
A0A1E1W163 I4DMH9 A0A194QKH9 A0A194PDJ6 A0A212F2X0 R4WSD0 D2A0T6 A0A023F8X0 A0A170XZ91 A0A224XS00 A0A1B6FPH7 A0A1B6K7G7 A0A2J7R8G9 A0A1B6KAI8 A0A0V0G2T5 A0A2P8YXN4 A0A182Y680 R4G4I3 A0A182W8P5 A0A182RWP2 A0A182NP04 U5ERS3 A0A0T6AU61 A0A182K1N5 A0A182X2G1 A0A182VH14 A0A182PVR2 Q7PMF2 A0A182HR72 Q179Z6 A0A0A9W715 A0A1B0CM87 Q1HRR4 A0A1L8DTN5 K7J126 A0A2M4BT50 A0A2M4A5N8 W5JGB6 A0A2M3ZEY0 A0A2M3Z0Z6 T1DU17 A0A2M4A5I5 A0A1Y1MRH2 A0A2M4CTQ5 A0A182J8L4 A0A182MDR5 A0A0C9QLB5 A0A2M4CFQ7 A0A0M3QTH1 A0A088AT27 A0A182TIU8 E2C774 A0A158P2D2 A0A1S3DR32 A0A182KZ67 A0A023EPV7 A0A026WE28 A0A2A3EG61 B0WEC9 A0A1Q3FBV3 A0A336M6Y7 E9IEY8 Q8SY53 B4Q6P9 B4HYI1 A0A232FNB6 B4MTJ2 W8CAX0 B3N736 A0A034V1T9 A0A0K8TZJ0 A0A0A1X2D3 Q29MN1 B4G903 A0A3B0K8F3 A0A1W4V7V4 B4KJM7 A0A151WTL0 B4NWV7 A0A182F357 E2AY50 B3MPG6 F4WUP4 B4LSH5 A0A0P6GR00 A0A195E505 A0A1J1HLU4 A0A195BTB2 A0A182GRF3 E9GTV6 F8TGY1 A0A1L8EH06 A0A336MBG5 D3TMZ0
A0A1E1W163 I4DMH9 A0A194QKH9 A0A194PDJ6 A0A212F2X0 R4WSD0 D2A0T6 A0A023F8X0 A0A170XZ91 A0A224XS00 A0A1B6FPH7 A0A1B6K7G7 A0A2J7R8G9 A0A1B6KAI8 A0A0V0G2T5 A0A2P8YXN4 A0A182Y680 R4G4I3 A0A182W8P5 A0A182RWP2 A0A182NP04 U5ERS3 A0A0T6AU61 A0A182K1N5 A0A182X2G1 A0A182VH14 A0A182PVR2 Q7PMF2 A0A182HR72 Q179Z6 A0A0A9W715 A0A1B0CM87 Q1HRR4 A0A1L8DTN5 K7J126 A0A2M4BT50 A0A2M4A5N8 W5JGB6 A0A2M3ZEY0 A0A2M3Z0Z6 T1DU17 A0A2M4A5I5 A0A1Y1MRH2 A0A2M4CTQ5 A0A182J8L4 A0A182MDR5 A0A0C9QLB5 A0A2M4CFQ7 A0A0M3QTH1 A0A088AT27 A0A182TIU8 E2C774 A0A158P2D2 A0A1S3DR32 A0A182KZ67 A0A023EPV7 A0A026WE28 A0A2A3EG61 B0WEC9 A0A1Q3FBV3 A0A336M6Y7 E9IEY8 Q8SY53 B4Q6P9 B4HYI1 A0A232FNB6 B4MTJ2 W8CAX0 B3N736 A0A034V1T9 A0A0K8TZJ0 A0A0A1X2D3 Q29MN1 B4G903 A0A3B0K8F3 A0A1W4V7V4 B4KJM7 A0A151WTL0 B4NWV7 A0A182F357 E2AY50 B3MPG6 F4WUP4 B4LSH5 A0A0P6GR00 A0A195E505 A0A1J1HLU4 A0A195BTB2 A0A182GRF3 E9GTV6 F8TGY1 A0A1L8EH06 A0A336MBG5 D3TMZ0
Pubmed
19121390
23622113
22651552
26354079
22118469
23691247
+ More
18362917 19820115 25474469 29403074 25244985 12364791 17510324 25401762 26823975 17204158 20075255 20920257 23761445 28004739 20798317 21347285 20966253 24945155 24508170 21282665 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 22936249 28648823 24495485 25348373 25830018 15632085 17550304 18057021 21719571 26483478 21292972 21668550 20353571
18362917 19820115 25474469 29403074 25244985 12364791 17510324 25401762 26823975 17204158 20075255 20920257 23761445 28004739 20798317 21347285 20966253 24945155 24508170 21282665 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 22936249 28648823 24495485 25348373 25830018 15632085 17550304 18057021 21719571 26483478 21292972 21668550 20353571
EMBL
BABH01001468
BABH01001469
BABH01001470
BABH01001471
RSAL01000017
RVE52906.1
+ More
ODYU01001899 SOQ38755.1 NWSH01000168 PCG78833.1 GAIX01004615 JAA87945.1 AK401629 BAM18251.1 GDQN01010314 JAT80740.1 AK402497 BAM19119.1 KQ461198 KPJ06073.1 KQ459606 KPI91083.1 AGBW02010668 OWR48077.1 AK417607 BAN20822.1 KQ971338 EFA01636.1 GBBI01000835 JAC17877.1 GEMB01003868 JAR99386.1 GFTR01005114 JAW11312.1 GECZ01017663 JAS52106.1 GECU01000331 JAT07376.1 NEVH01006721 PNF37128.1 GEBQ01031519 JAT08458.1 GECL01003695 JAP02429.1 PYGN01000298 PSN49006.1 ACPB03019965 GAHY01001335 JAA76175.1 GANO01002689 JAB57182.1 LJIG01022810 KRT78601.1 AAAB01008980 EAA13927.5 APCN01001654 CH477342 EAT43075.1 GBHO01040403 GDHC01006388 JAG03201.1 JAQ12241.1 AJWK01018369 DQ440030 ABF18063.1 GFDF01004261 JAV09823.1 GGFJ01007125 MBW56266.1 GGFK01002740 MBW36061.1 ADMH02001597 ETN61920.1 GGFM01006312 MBW27063.1 GGFM01001451 MBW22202.1 GAMD01000559 JAB01032.1 GGFK01002746 MBW36067.1 GEZM01023579 JAV88293.1 GGFL01004558 MBW68736.1 AXCM01000350 GBYB01001394 GBYB01001397 JAG71161.1 JAG71164.1 GGFL01000002 MBW64180.1 CP012523 ALC38903.1 GL453340 EFN76187.1 ADTU01007116 GAPW01002543 JAC11055.1 KK107250 EZA54360.1 KZ288254 PBC30763.1 DS231907 EDS45489.1 GFDL01009984 JAV25061.1 UFQT01000478 SSX24649.1 GL762730 EFZ20865.1 AE014134 AY075331 AAF52636.2 AAL68198.1 CM000361 CM002910 EDX04208.1 KMY89034.1 CH480818 EDW52111.1 NNAY01000009 OXU32073.1 CH963852 EDW75431.1 GAMC01005341 GAMC01005339 JAC01215.1 CH954177 EDV59332.1 GAKP01023430 JAC35528.1 GDHF01032626 GDHF01009869 JAI19688.1 JAI42445.1 GBXI01008783 JAD05509.1 CH379060 EAL33662.1 CH479180 EDW28833.1 OUUW01000004 SPP79808.1 CH933807 EDW11472.1 KQ982753 KYQ51174.1 CM000157 EDW88487.1 GL443762 EFN61646.1 CH902620 EDV31262.1 KPU73288.1 GL888375 EGI62041.1 CH940649 EDW64797.1 GDIP01117145 GDIQ01029894 GDIQ01002793 LRGB01002384 JAL86569.1 JAN64843.1 KZS07981.1 KQ979657 KYN19982.1 CVRI01000008 CRK88514.1 KQ976408 KYM91176.1 JXUM01016218 KQ560452 KXJ82352.1 GL732565 EFX77074.1 HQ896857 HQ896858 AEI98820.1 GFDG01000802 JAV17997.1 UFQT01000091 UFQT01000704 SSX19570.1 SSX26671.1 CCAG010021443 EZ422792 ADD19068.1
ODYU01001899 SOQ38755.1 NWSH01000168 PCG78833.1 GAIX01004615 JAA87945.1 AK401629 BAM18251.1 GDQN01010314 JAT80740.1 AK402497 BAM19119.1 KQ461198 KPJ06073.1 KQ459606 KPI91083.1 AGBW02010668 OWR48077.1 AK417607 BAN20822.1 KQ971338 EFA01636.1 GBBI01000835 JAC17877.1 GEMB01003868 JAR99386.1 GFTR01005114 JAW11312.1 GECZ01017663 JAS52106.1 GECU01000331 JAT07376.1 NEVH01006721 PNF37128.1 GEBQ01031519 JAT08458.1 GECL01003695 JAP02429.1 PYGN01000298 PSN49006.1 ACPB03019965 GAHY01001335 JAA76175.1 GANO01002689 JAB57182.1 LJIG01022810 KRT78601.1 AAAB01008980 EAA13927.5 APCN01001654 CH477342 EAT43075.1 GBHO01040403 GDHC01006388 JAG03201.1 JAQ12241.1 AJWK01018369 DQ440030 ABF18063.1 GFDF01004261 JAV09823.1 GGFJ01007125 MBW56266.1 GGFK01002740 MBW36061.1 ADMH02001597 ETN61920.1 GGFM01006312 MBW27063.1 GGFM01001451 MBW22202.1 GAMD01000559 JAB01032.1 GGFK01002746 MBW36067.1 GEZM01023579 JAV88293.1 GGFL01004558 MBW68736.1 AXCM01000350 GBYB01001394 GBYB01001397 JAG71161.1 JAG71164.1 GGFL01000002 MBW64180.1 CP012523 ALC38903.1 GL453340 EFN76187.1 ADTU01007116 GAPW01002543 JAC11055.1 KK107250 EZA54360.1 KZ288254 PBC30763.1 DS231907 EDS45489.1 GFDL01009984 JAV25061.1 UFQT01000478 SSX24649.1 GL762730 EFZ20865.1 AE014134 AY075331 AAF52636.2 AAL68198.1 CM000361 CM002910 EDX04208.1 KMY89034.1 CH480818 EDW52111.1 NNAY01000009 OXU32073.1 CH963852 EDW75431.1 GAMC01005341 GAMC01005339 JAC01215.1 CH954177 EDV59332.1 GAKP01023430 JAC35528.1 GDHF01032626 GDHF01009869 JAI19688.1 JAI42445.1 GBXI01008783 JAD05509.1 CH379060 EAL33662.1 CH479180 EDW28833.1 OUUW01000004 SPP79808.1 CH933807 EDW11472.1 KQ982753 KYQ51174.1 CM000157 EDW88487.1 GL443762 EFN61646.1 CH902620 EDV31262.1 KPU73288.1 GL888375 EGI62041.1 CH940649 EDW64797.1 GDIP01117145 GDIQ01029894 GDIQ01002793 LRGB01002384 JAL86569.1 JAN64843.1 KZS07981.1 KQ979657 KYN19982.1 CVRI01000008 CRK88514.1 KQ976408 KYM91176.1 JXUM01016218 KQ560452 KXJ82352.1 GL732565 EFX77074.1 HQ896857 HQ896858 AEI98820.1 GFDG01000802 JAV17997.1 UFQT01000091 UFQT01000704 SSX19570.1 SSX26671.1 CCAG010021443 EZ422792 ADD19068.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053240
UP000053268
UP000007151
+ More
UP000007266 UP000235965 UP000245037 UP000076408 UP000015103 UP000075920 UP000075900 UP000075884 UP000075881 UP000076407 UP000075903 UP000075885 UP000007062 UP000075840 UP000008820 UP000092461 UP000002358 UP000000673 UP000075880 UP000075883 UP000092553 UP000005203 UP000075902 UP000008237 UP000005205 UP000079169 UP000075882 UP000053097 UP000242457 UP000002320 UP000000803 UP000000304 UP000001292 UP000215335 UP000007798 UP000008711 UP000001819 UP000008744 UP000268350 UP000192221 UP000009192 UP000075809 UP000002282 UP000069272 UP000000311 UP000007801 UP000007755 UP000008792 UP000076858 UP000078492 UP000183832 UP000078540 UP000069940 UP000249989 UP000000305 UP000092444
UP000007266 UP000235965 UP000245037 UP000076408 UP000015103 UP000075920 UP000075900 UP000075884 UP000075881 UP000076407 UP000075903 UP000075885 UP000007062 UP000075840 UP000008820 UP000092461 UP000002358 UP000000673 UP000075880 UP000075883 UP000092553 UP000005203 UP000075902 UP000008237 UP000005205 UP000079169 UP000075882 UP000053097 UP000242457 UP000002320 UP000000803 UP000000304 UP000001292 UP000215335 UP000007798 UP000008711 UP000001819 UP000008744 UP000268350 UP000192221 UP000009192 UP000075809 UP000002282 UP000069272 UP000000311 UP000007801 UP000007755 UP000008792 UP000076858 UP000078492 UP000183832 UP000078540 UP000069940 UP000249989 UP000000305 UP000092444
Interpro
ProteinModelPortal
H9IVM3
A0A3S2NJZ2
A0A2H1VD56
A0A2A4K4Y6
S4PKA2
I4DK11
+ More
A0A1E1W163 I4DMH9 A0A194QKH9 A0A194PDJ6 A0A212F2X0 R4WSD0 D2A0T6 A0A023F8X0 A0A170XZ91 A0A224XS00 A0A1B6FPH7 A0A1B6K7G7 A0A2J7R8G9 A0A1B6KAI8 A0A0V0G2T5 A0A2P8YXN4 A0A182Y680 R4G4I3 A0A182W8P5 A0A182RWP2 A0A182NP04 U5ERS3 A0A0T6AU61 A0A182K1N5 A0A182X2G1 A0A182VH14 A0A182PVR2 Q7PMF2 A0A182HR72 Q179Z6 A0A0A9W715 A0A1B0CM87 Q1HRR4 A0A1L8DTN5 K7J126 A0A2M4BT50 A0A2M4A5N8 W5JGB6 A0A2M3ZEY0 A0A2M3Z0Z6 T1DU17 A0A2M4A5I5 A0A1Y1MRH2 A0A2M4CTQ5 A0A182J8L4 A0A182MDR5 A0A0C9QLB5 A0A2M4CFQ7 A0A0M3QTH1 A0A088AT27 A0A182TIU8 E2C774 A0A158P2D2 A0A1S3DR32 A0A182KZ67 A0A023EPV7 A0A026WE28 A0A2A3EG61 B0WEC9 A0A1Q3FBV3 A0A336M6Y7 E9IEY8 Q8SY53 B4Q6P9 B4HYI1 A0A232FNB6 B4MTJ2 W8CAX0 B3N736 A0A034V1T9 A0A0K8TZJ0 A0A0A1X2D3 Q29MN1 B4G903 A0A3B0K8F3 A0A1W4V7V4 B4KJM7 A0A151WTL0 B4NWV7 A0A182F357 E2AY50 B3MPG6 F4WUP4 B4LSH5 A0A0P6GR00 A0A195E505 A0A1J1HLU4 A0A195BTB2 A0A182GRF3 E9GTV6 F8TGY1 A0A1L8EH06 A0A336MBG5 D3TMZ0
A0A1E1W163 I4DMH9 A0A194QKH9 A0A194PDJ6 A0A212F2X0 R4WSD0 D2A0T6 A0A023F8X0 A0A170XZ91 A0A224XS00 A0A1B6FPH7 A0A1B6K7G7 A0A2J7R8G9 A0A1B6KAI8 A0A0V0G2T5 A0A2P8YXN4 A0A182Y680 R4G4I3 A0A182W8P5 A0A182RWP2 A0A182NP04 U5ERS3 A0A0T6AU61 A0A182K1N5 A0A182X2G1 A0A182VH14 A0A182PVR2 Q7PMF2 A0A182HR72 Q179Z6 A0A0A9W715 A0A1B0CM87 Q1HRR4 A0A1L8DTN5 K7J126 A0A2M4BT50 A0A2M4A5N8 W5JGB6 A0A2M3ZEY0 A0A2M3Z0Z6 T1DU17 A0A2M4A5I5 A0A1Y1MRH2 A0A2M4CTQ5 A0A182J8L4 A0A182MDR5 A0A0C9QLB5 A0A2M4CFQ7 A0A0M3QTH1 A0A088AT27 A0A182TIU8 E2C774 A0A158P2D2 A0A1S3DR32 A0A182KZ67 A0A023EPV7 A0A026WE28 A0A2A3EG61 B0WEC9 A0A1Q3FBV3 A0A336M6Y7 E9IEY8 Q8SY53 B4Q6P9 B4HYI1 A0A232FNB6 B4MTJ2 W8CAX0 B3N736 A0A034V1T9 A0A0K8TZJ0 A0A0A1X2D3 Q29MN1 B4G903 A0A3B0K8F3 A0A1W4V7V4 B4KJM7 A0A151WTL0 B4NWV7 A0A182F357 E2AY50 B3MPG6 F4WUP4 B4LSH5 A0A0P6GR00 A0A195E505 A0A1J1HLU4 A0A195BTB2 A0A182GRF3 E9GTV6 F8TGY1 A0A1L8EH06 A0A336MBG5 D3TMZ0
PDB
4M90
E-value=3.52378e-49,
Score=491
Ontologies
PATHWAY
GO
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 20,
Likelihood: 0.903454
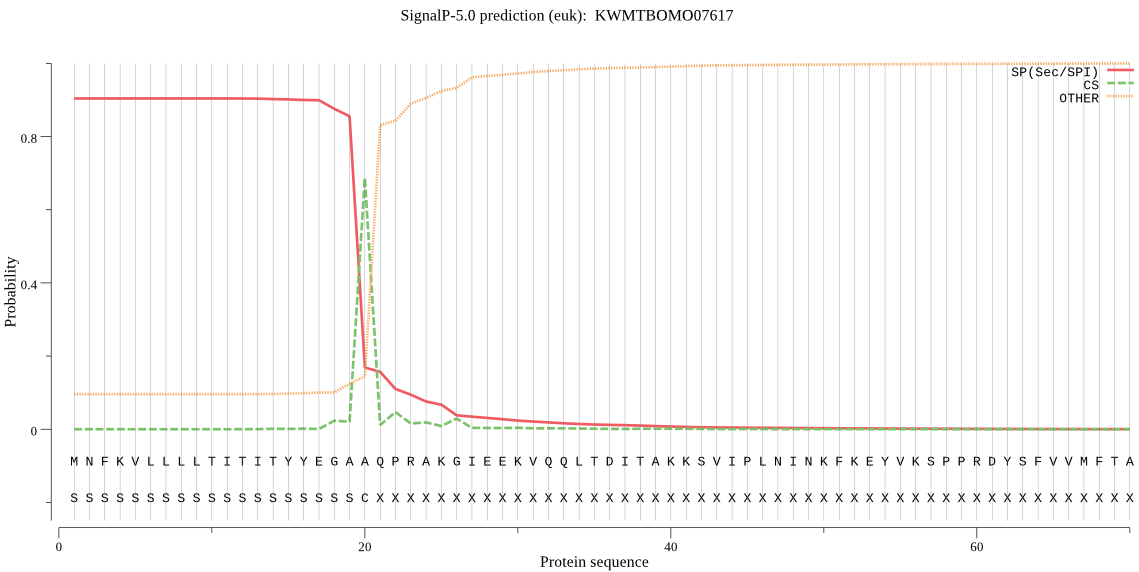
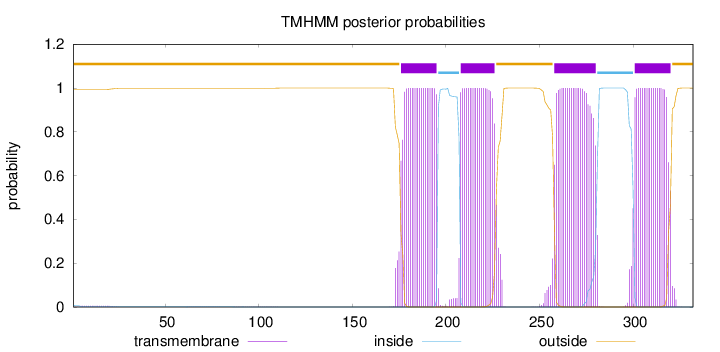
Length:
332
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
83.45755
Exp number, first 60 AAs:
0.10425
Total prob of N-in:
0.00659
outside
1 - 175
TMhelix
176 - 195
inside
196 - 207
TMhelix
208 - 226
outside
227 - 257
TMhelix
258 - 280
inside
281 - 300
TMhelix
301 - 320
outside
321 - 332
Population Genetic Test Statistics
Pi
20.933183
Theta
28.200749
Tajima's D
-1.154683
CLR
117.541804
CSRT
0.108644567771611
Interpretation
Uncertain
