Gene
KWMTBOMO07604
Pre Gene Modal
BGIBMGA013711
Annotation
PREDICTED:_organic_cation_transporter_protein-like_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.966
Sequence
CDS
ATGGCGTCAACGGAAATTGAATTCGAAGATATAATAGAAAAATTCAAACTTTTCTCTCCTTATCATTTAAAACTGGTTGTTCTGCTTGCGTTCGTCAGTTTTATTGACGCTTGGCATCGTGCTAATTACATTATGGTCGTCGAAGACGTTGATTACTGGTGTAAATACGAATGTAATGAAAGCCAGTTTCTTAAAGTTATTCGTAACGAAACTAATCTAGACAGATGTTTGAAGTACTCATTTACTGACAATTGCGCTTTCGATGACCTTAAAGAAAGAAGCATCGAAAACTGCCATGACTGGACCTATGAGAAACCTGACAGTTTTATGGGTGAATTCAAACTGGGATGTCAGGATTGGAAAAGAACGTTAGTGGGGACTGTCCACAGCATGGGTCTTTTGATCGGCTTACTCATTATAGGACCCCTGTCTGATAAATACGGCCGAAAAGTCATATTTATACTCTGCGGAATAGTGGGTGGCATTGTTGGATTAGGCAAAAGCATAACGGCGTGGTACTGGATTTATGTAACACTGGAGTTTGTTCAAGTCTTAATCGGAACTCCGTTTTCGGCAGCTTTTACACTGGGGATAGAGATGGTACCGAAACGTAACCGCCTACGTTTTCTTGGCCTCGTTTCATCTTTCTTCGGTGTCGGCGACATCACCCTCGCTCTACTAGCTTGGTCAGTACCGTACTGGAGGAACCTCTTGAGAGTCCTTTACGCACCATCTTTCATCTTCTTAATTTACATCTTCGCAATGGACGAAAGCATTCGATGGTTGTTGATAAAGGGAAGAAAAGAAGAGGCAATTAATATCATACAGAAAGCAGCTAAAGTGAATAAAATCCACATCAGCAATGAAGATCTGCATCGTATGAAAAGCGAGAAAACGTCAAAAAACACGGGCCTTCTGGACCTGTTGAAGATAACGATGAAATCGAGGAAGTTGTTGCTGAGGACTCTGATGTGTGTTTTGATGTGGATGACGTCATCTTTCAGTGTTTATACTCTATTAATCAATTCCGTCAGTTTGGCTGGAAATAAATACTTGAATTACGGTGCTGTTGCTTGTGCTAGTTCAATGGCAGTTCTATCGGGAGGTTTTTTAATGGAAAATCTCAAACGAAGAGTTCCTCTTATCACCGGGATGGGTATGACTGGTCTTATTTTTATAATTCAGTCATTGGTTCCTAAAGAATATTCTTTGTTGGCGGCCGTGTTCTTTTTTATCGGCAAATACTTCTCATCACTGGCATTTTTATTTATTTATTTGTATACTTCAGAATTATTCCCAACGTACACGAGAAATACTATGCATGCATTTTGTTCTTCTGTCGGTAGAATCGGATCCATTTTAGCCCCTCAAACTCCACTGCTGACACAATATTGGTCGGGCATTCCTTCCATGCTAGTGGGGTCCCTGTCTTTGATCACAGCCATCGCTATCTTTCTTGTTCCTGACACATCGGAAGACCCGCTACCAGACACGGTGGAAGAGGCTGAAGCTATCGGACACAAAAAGAACAATATCTCTAAATCAAGAGTAACAATAATTTCATCATGTTGTTAA
Protein
MASTEIEFEDIIEKFKLFSPYHLKLVVLLAFVSFIDAWHRANYIMVVEDVDYWCKYECNESQFLKVIRNETNLDRCLKYSFTDNCAFDDLKERSIENCHDWTYEKPDSFMGEFKLGCQDWKRTLVGTVHSMGLLIGLLIIGPLSDKYGRKVIFILCGIVGGIVGLGKSITAWYWIYVTLEFVQVLIGTPFSAAFTLGIEMVPKRNRLRFLGLVSSFFGVGDITLALLAWSVPYWRNLLRVLYAPSFIFLIYIFAMDESIRWLLIKGRKEEAINIIQKAAKVNKIHISNEDLHRMKSEKTSKNTGLLDLLKITMKSRKLLLRTLMCVLMWMTSSFSVYTLLINSVSLAGNKYLNYGAVACASSMAVLSGGFLMENLKRRVPLITGMGMTGLIFIIQSLVPKEYSLLAAVFFFIGKYFSSLAFLFIYLYTSELFPTYTRNTMHAFCSSVGRIGSILAPQTPLLTQYWSGIPSMLVGSLSLITAIAIFLVPDTSEDPLPDTVEEAEAIGHKKNNISKSRVTIISSCC
Summary
Uniprot
A0A2A4K4I8
A0A2A4K3J3
A0A2A4K592
H9JVZ5
A0A2H1WTZ6
A0A2H1VFZ4
+ More
A0A212EH35 S4P1Y2 A0A2A4JPL4 A0A2W1BM98 A0A212F2F2 A0A2A4J3F4 A0A2H1VQS6 A0A194PCX0 A0A194RH24 H9JVZ7 A0A194PCQ2 A0A212EWR7 A0A3S2NA23 A0A212EJ20 A0A0N0P9G0 A0A3S2NIQ2 A0A3S2M6B8 A0A194QDP3 A0A0L7LVH1 A0A0L7KUG4 A0A212FPC5 A0A2H1VA76 A0A2W1BRJ1 A0A2A4K5R4 A0A2H1W8C3 A0A2H1V9B3 A0A194PT22 A0A194PSH8 A0A2A4K7D4 A0A2A4K7E3 A0A0N1I6I7 A0A194QC91 A0A2H1V1L3 A0A2W1BUD3 A0A2A4J6J0 A0A3S2NDN7 H9IWG7 A0A194RIS2 A0A2A4J5Q3 A0A194REW9 A0A212F2Q3 A0A2A4K6M9 A0A3S2L3J0 A0A2A4K5P1 A0A194PJI6 A0A2A4J455 A0A067RSY2 A0A084VS25 A0A212F158 A0A212F848 A0A212EJ24 A0A194Q2K7 A0A0K8VUC5 A0A3S2TCP3 A0A0K8VEY5 A0A0K8UFN6 A0A0L7LKI8 A0A2H1V9G0 A0A0K8UAY5 A0A212F141 U5EVF0 A0A194RGD2 H9IVN2 A0A2H1VFS1 A0A194RER3 A0A182Y1J2 A0A1Y1KD84 A0A2A4JC88 A0A182FXJ6 A0A2A4K5Q2 A0A067RGL9 A0A182J645 A0A194PT27 A0A158P2C5 A0A212EIZ7 A0A194PXP4 B0XDF1 A0A2W1BMH3 A0A194RE10 H9JI49 A0A182M3B6 A0A194QC06 A0A034WIX2 A0A0V0GBF3 A0A182TM67 A0A194RF43 A0A182VDY0 A0A154PCE9 F4WUN7 A0A195E462 A0A182MYV4
A0A212EH35 S4P1Y2 A0A2A4JPL4 A0A2W1BM98 A0A212F2F2 A0A2A4J3F4 A0A2H1VQS6 A0A194PCX0 A0A194RH24 H9JVZ7 A0A194PCQ2 A0A212EWR7 A0A3S2NA23 A0A212EJ20 A0A0N0P9G0 A0A3S2NIQ2 A0A3S2M6B8 A0A194QDP3 A0A0L7LVH1 A0A0L7KUG4 A0A212FPC5 A0A2H1VA76 A0A2W1BRJ1 A0A2A4K5R4 A0A2H1W8C3 A0A2H1V9B3 A0A194PT22 A0A194PSH8 A0A2A4K7D4 A0A2A4K7E3 A0A0N1I6I7 A0A194QC91 A0A2H1V1L3 A0A2W1BUD3 A0A2A4J6J0 A0A3S2NDN7 H9IWG7 A0A194RIS2 A0A2A4J5Q3 A0A194REW9 A0A212F2Q3 A0A2A4K6M9 A0A3S2L3J0 A0A2A4K5P1 A0A194PJI6 A0A2A4J455 A0A067RSY2 A0A084VS25 A0A212F158 A0A212F848 A0A212EJ24 A0A194Q2K7 A0A0K8VUC5 A0A3S2TCP3 A0A0K8VEY5 A0A0K8UFN6 A0A0L7LKI8 A0A2H1V9G0 A0A0K8UAY5 A0A212F141 U5EVF0 A0A194RGD2 H9IVN2 A0A2H1VFS1 A0A194RER3 A0A182Y1J2 A0A1Y1KD84 A0A2A4JC88 A0A182FXJ6 A0A2A4K5Q2 A0A067RGL9 A0A182J645 A0A194PT27 A0A158P2C5 A0A212EIZ7 A0A194PXP4 B0XDF1 A0A2W1BMH3 A0A194RE10 H9JI49 A0A182M3B6 A0A194QC06 A0A034WIX2 A0A0V0GBF3 A0A182TM67 A0A194RF43 A0A182VDY0 A0A154PCE9 F4WUN7 A0A195E462 A0A182MYV4
Pubmed
EMBL
NWSH01000168
PCG78824.1
PCG78831.1
PCG78830.1
BABH01036474
ODYU01010990
+ More
SOQ56446.1 ODYU01002336 SOQ39701.1 AGBW02014973 OWR40799.1 GAIX01012265 JAA80295.1 NWSH01000834 PCG73941.1 KZ149965 PZC76202.1 AGBW02010753 OWR47904.1 NWSH01003619 PCG66044.1 ODYU01003881 SOQ43193.1 KQ459606 KPI91072.1 KQ460205 KPJ17123.1 BABH01036481 KPI91071.1 AGBW02011950 OWR45874.1 RSAL01000360 RVE42197.1 AGBW02014542 OWR41475.1 KQ459449 KPJ00906.1 RSAL01000974 RVE41008.1 RSAL01000027 RVE51971.1 KQ459193 KPJ03085.1 JTDY01000022 KOB79379.1 JTDY01005496 KOB66937.1 AGBW02003000 OWR55596.1 ODYU01001220 SOQ37134.1 KZ149964 PZC76264.1 NWSH01000100 PCG79571.1 ODYU01006920 SOQ49196.1 ODYU01001366 SOQ37440.1 KQ459595 KPI95914.1 KPI95923.1 PCG79560.1 PCG79570.1 KQ461184 KPJ07510.1 KPJ03082.1 ODYU01000079 SOQ34232.1 PZC76256.1 NWSH01002815 PCG67489.1 RSAL01000186 RVE44804.1 BABH01006647 KQ460325 KPJ15841.1 NWSH01003151 PCG66854.1 KPJ15840.1 AGBW02010692 OWR48028.1 PCG79564.1 RVE44802.1 PCG79565.1 KPI91250.1 PCG66855.1 KK852482 KDR22929.1 ATLV01015792 KE525036 KFB40769.1 AGBW02010973 OWR47451.1 AGBW02009797 OWR49901.1 OWR41479.1 KQ459562 KPI99797.1 GDHF01009833 JAI42481.1 RVE42200.1 GDHF01014878 JAI37436.1 GDHF01026931 GDHF01013660 GDHF01009296 JAI25383.1 JAI38654.1 JAI43018.1 JTDY01000842 KOB75696.1 SOQ37441.1 GDHF01028477 JAI23837.1 OWR47447.1 GANO01001906 JAB57965.1 KPJ16878.1 BABH01039623 ODYU01002328 SOQ39679.1 KQ460313 KPJ16067.1 GEZM01087818 JAV58458.1 NWSH01002161 PCG69013.1 PCG79557.1 KDR22927.1 KPI95919.1 ADTU01007112 AGBW02014543 OWR41473.1 KPI95920.1 DS232756 EDS45429.1 PZC76259.1 KPJ16068.1 BABH01041119 BABH01041120 AXCM01003135 KPJ03083.1 GAKP01005259 JAC53693.1 GECL01000840 JAP05284.1 KPJ16069.1 KQ434870 KZC09576.1 GL888375 EGI62034.1 KQ979657 KYN19975.1
SOQ56446.1 ODYU01002336 SOQ39701.1 AGBW02014973 OWR40799.1 GAIX01012265 JAA80295.1 NWSH01000834 PCG73941.1 KZ149965 PZC76202.1 AGBW02010753 OWR47904.1 NWSH01003619 PCG66044.1 ODYU01003881 SOQ43193.1 KQ459606 KPI91072.1 KQ460205 KPJ17123.1 BABH01036481 KPI91071.1 AGBW02011950 OWR45874.1 RSAL01000360 RVE42197.1 AGBW02014542 OWR41475.1 KQ459449 KPJ00906.1 RSAL01000974 RVE41008.1 RSAL01000027 RVE51971.1 KQ459193 KPJ03085.1 JTDY01000022 KOB79379.1 JTDY01005496 KOB66937.1 AGBW02003000 OWR55596.1 ODYU01001220 SOQ37134.1 KZ149964 PZC76264.1 NWSH01000100 PCG79571.1 ODYU01006920 SOQ49196.1 ODYU01001366 SOQ37440.1 KQ459595 KPI95914.1 KPI95923.1 PCG79560.1 PCG79570.1 KQ461184 KPJ07510.1 KPJ03082.1 ODYU01000079 SOQ34232.1 PZC76256.1 NWSH01002815 PCG67489.1 RSAL01000186 RVE44804.1 BABH01006647 KQ460325 KPJ15841.1 NWSH01003151 PCG66854.1 KPJ15840.1 AGBW02010692 OWR48028.1 PCG79564.1 RVE44802.1 PCG79565.1 KPI91250.1 PCG66855.1 KK852482 KDR22929.1 ATLV01015792 KE525036 KFB40769.1 AGBW02010973 OWR47451.1 AGBW02009797 OWR49901.1 OWR41479.1 KQ459562 KPI99797.1 GDHF01009833 JAI42481.1 RVE42200.1 GDHF01014878 JAI37436.1 GDHF01026931 GDHF01013660 GDHF01009296 JAI25383.1 JAI38654.1 JAI43018.1 JTDY01000842 KOB75696.1 SOQ37441.1 GDHF01028477 JAI23837.1 OWR47447.1 GANO01001906 JAB57965.1 KPJ16878.1 BABH01039623 ODYU01002328 SOQ39679.1 KQ460313 KPJ16067.1 GEZM01087818 JAV58458.1 NWSH01002161 PCG69013.1 PCG79557.1 KDR22927.1 KPI95919.1 ADTU01007112 AGBW02014543 OWR41473.1 KPI95920.1 DS232756 EDS45429.1 PZC76259.1 KPJ16068.1 BABH01041119 BABH01041120 AXCM01003135 KPJ03083.1 GAKP01005259 JAC53693.1 GECL01000840 JAP05284.1 KPJ16069.1 KQ434870 KZC09576.1 GL888375 EGI62034.1 KQ979657 KYN19975.1
Proteomes
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
A0A2A4K4I8
A0A2A4K3J3
A0A2A4K592
H9JVZ5
A0A2H1WTZ6
A0A2H1VFZ4
+ More
A0A212EH35 S4P1Y2 A0A2A4JPL4 A0A2W1BM98 A0A212F2F2 A0A2A4J3F4 A0A2H1VQS6 A0A194PCX0 A0A194RH24 H9JVZ7 A0A194PCQ2 A0A212EWR7 A0A3S2NA23 A0A212EJ20 A0A0N0P9G0 A0A3S2NIQ2 A0A3S2M6B8 A0A194QDP3 A0A0L7LVH1 A0A0L7KUG4 A0A212FPC5 A0A2H1VA76 A0A2W1BRJ1 A0A2A4K5R4 A0A2H1W8C3 A0A2H1V9B3 A0A194PT22 A0A194PSH8 A0A2A4K7D4 A0A2A4K7E3 A0A0N1I6I7 A0A194QC91 A0A2H1V1L3 A0A2W1BUD3 A0A2A4J6J0 A0A3S2NDN7 H9IWG7 A0A194RIS2 A0A2A4J5Q3 A0A194REW9 A0A212F2Q3 A0A2A4K6M9 A0A3S2L3J0 A0A2A4K5P1 A0A194PJI6 A0A2A4J455 A0A067RSY2 A0A084VS25 A0A212F158 A0A212F848 A0A212EJ24 A0A194Q2K7 A0A0K8VUC5 A0A3S2TCP3 A0A0K8VEY5 A0A0K8UFN6 A0A0L7LKI8 A0A2H1V9G0 A0A0K8UAY5 A0A212F141 U5EVF0 A0A194RGD2 H9IVN2 A0A2H1VFS1 A0A194RER3 A0A182Y1J2 A0A1Y1KD84 A0A2A4JC88 A0A182FXJ6 A0A2A4K5Q2 A0A067RGL9 A0A182J645 A0A194PT27 A0A158P2C5 A0A212EIZ7 A0A194PXP4 B0XDF1 A0A2W1BMH3 A0A194RE10 H9JI49 A0A182M3B6 A0A194QC06 A0A034WIX2 A0A0V0GBF3 A0A182TM67 A0A194RF43 A0A182VDY0 A0A154PCE9 F4WUN7 A0A195E462 A0A182MYV4
A0A212EH35 S4P1Y2 A0A2A4JPL4 A0A2W1BM98 A0A212F2F2 A0A2A4J3F4 A0A2H1VQS6 A0A194PCX0 A0A194RH24 H9JVZ7 A0A194PCQ2 A0A212EWR7 A0A3S2NA23 A0A212EJ20 A0A0N0P9G0 A0A3S2NIQ2 A0A3S2M6B8 A0A194QDP3 A0A0L7LVH1 A0A0L7KUG4 A0A212FPC5 A0A2H1VA76 A0A2W1BRJ1 A0A2A4K5R4 A0A2H1W8C3 A0A2H1V9B3 A0A194PT22 A0A194PSH8 A0A2A4K7D4 A0A2A4K7E3 A0A0N1I6I7 A0A194QC91 A0A2H1V1L3 A0A2W1BUD3 A0A2A4J6J0 A0A3S2NDN7 H9IWG7 A0A194RIS2 A0A2A4J5Q3 A0A194REW9 A0A212F2Q3 A0A2A4K6M9 A0A3S2L3J0 A0A2A4K5P1 A0A194PJI6 A0A2A4J455 A0A067RSY2 A0A084VS25 A0A212F158 A0A212F848 A0A212EJ24 A0A194Q2K7 A0A0K8VUC5 A0A3S2TCP3 A0A0K8VEY5 A0A0K8UFN6 A0A0L7LKI8 A0A2H1V9G0 A0A0K8UAY5 A0A212F141 U5EVF0 A0A194RGD2 H9IVN2 A0A2H1VFS1 A0A194RER3 A0A182Y1J2 A0A1Y1KD84 A0A2A4JC88 A0A182FXJ6 A0A2A4K5Q2 A0A067RGL9 A0A182J645 A0A194PT27 A0A158P2C5 A0A212EIZ7 A0A194PXP4 B0XDF1 A0A2W1BMH3 A0A194RE10 H9JI49 A0A182M3B6 A0A194QC06 A0A034WIX2 A0A0V0GBF3 A0A182TM67 A0A194RF43 A0A182VDY0 A0A154PCE9 F4WUN7 A0A195E462 A0A182MYV4
PDB
4LDS
E-value=0.00103751,
Score=101
Ontologies
GO
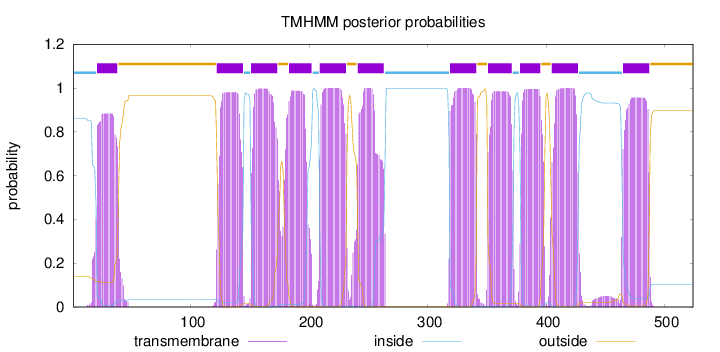
Topology
Length:
524
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
233.30597
Exp number, first 60 AAs:
17.71751
Total prob of N-in:
0.86070
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 38
outside
39 - 121
TMhelix
122 - 144
inside
145 - 150
TMhelix
151 - 173
outside
174 - 182
TMhelix
183 - 202
inside
203 - 208
TMhelix
209 - 231
outside
232 - 240
TMhelix
241 - 263
inside
264 - 318
TMhelix
319 - 341
outside
342 - 350
TMhelix
351 - 371
inside
372 - 377
TMhelix
378 - 395
outside
396 - 404
TMhelix
405 - 427
inside
428 - 464
TMhelix
465 - 487
outside
488 - 524
Population Genetic Test Statistics
Pi
190.689573
Theta
158.615284
Tajima's D
0.378609
CLR
0.00399
CSRT
0.47797610119494
Interpretation
Uncertain
