Gene
KWMTBOMO07602
Pre Gene Modal
BGIBMGA013707
Annotation
Exocyst_complex_component_7_[Papilio_machaon]
Full name
Exocyst complex component 7
Alternative Name
Exocyst complex component Exo70
Location in the cell
Cytoplasmic Reliability : 1.059 Mitochondrial Reliability : 1.497 Nuclear Reliability : 1.134
Sequence
CDS
ATGTTTTGTTTCAACCGCGAGTGGGAGGAGTGCACGCGGCTGCAGCGCGGGTACAGCGTGCCCGACGCCGAGCTGCGGGAGGCCCTGAAGCGCGACAACAAGCAGGCGCTGCTGCCGCCCTACACGGCCTTCTACGACGCCGCCGCCGCGCTGCCCTTCTCCAAGAACCCCGACAAGTACGTCAAGTACACGCCGCTGCAGATCGCCACCCAGCTGGACGGGTACTTCGATGAGGCTGCTTAG
Protein
MFCFNREWEECTRLQRGYSVPDAELREALKRDNKQALLPPYTAFYDAAAALPFSKNPDKYVKYTPLQIATQLDGYFDEAA
Summary
Description
Component of the exocyst complex involved in the docking of exocytic vesicles with fusion sites on the plasma membrane.
Required for exocytosis. Thought to function in intracellular vesicle targeting and docking before SNARE complex formation.
Required for exocytosis. Thought to function in intracellular vesicle targeting and docking before SNARE complex formation.
Subunit
The exocyst complex is composed of Sec3/Exoc1, Sec5/Exoc2, Sec6/Exoc3, Sec8/Exoc4, Sec10/Exoc5, Sec15/Exoc6, Exo70/Exoc7 and Exo84/Exoc8.
Similarity
Belongs to the EXO70 family.
Keywords
Complete proteome
Exocytosis
Phosphoprotein
Protein transport
Reference proteome
Transport
Feature
chain Exocyst complex component 7
Uniprot
A0A194RH19
A0A194PEF4
A0A2A4IXA0
A0A2H1X227
A0A212F586
A0A3Q0JAX0
+ More
H9JVZ4 A0A0P4VT57 A0A1B6DEN9 A0A1B6HYJ6 A0A1B6GH39 A0A146LWS2 A0A2H8TXB5 J9JWJ5 A0A067QTF9 A0A1B6KMY7 E0VIX9 A0A1W4WV82 A0A2H8TVN7 A0A2J7Q1Y7 A0A2J7Q1X1 A0A293MLS7 A0A293N5A1 A0A0P5FQT5 A0A0P5PJK7 A0A0P6G3C2 A0A0P4XNI7 A0A0N8CJ02 A0A2R5LMB4 A0A0N8A8B2 E9G2G2 A0A131YL43 L7M7U3 A0A131XFQ1 A0A1E1X4Y2 V5IBQ0 A0A1W4WJ56 A0A2P6KMS5 A0A224YMM6 A0A087US49 D6WW45 B7P4G4 V5HQ58 U4TYQ4 N6UH50 A0A232F0I2 A0A2P2I6K8 A0A0A1X0B2 A0A0K8VY37 A0A034V746 B5DPB0 B4LDS3 B3M5I6 A0A0N0U3D4 A0A2A3E819 A0A088A2Q3 W8BLS5 B4IZE4 A0A1W4UES8 A0A0P4WFK6 A0A1I8PUR4 B4KYE1 A0A1B0DNY5 A0A1V9XU82 A0A3B0KAW7 A0A3B0KG25 A0A3R7QVN1 A0A1B0CUG9 A0A1A9UG92 A0A0L7R7B6 A0A1B0FPI7 A0A336MQ60 A0A336MIU8 C3Y9B1 A0A0K8TLX6 A0A1I8JVJ1 A0A1Q3FD21 A0A182X4R0 A0A182KAK4 A0A182U1X3 A0A182I4Q9 Q5TQS7 A0A1S3HN93 A0A1S3IE79 B0X634 A0A182KNB5 A0A182H9F6 A0A182PHS1 A0A226DDK3 D5AEM7 A0A182IQX8 A0A0J9RSV2 H1UUN4 B3NFS2 Q9VSJ8 B4PG27 A0A1J1HJL0 A0A1L8DVA6 B4N6D7 B4HJI4
H9JVZ4 A0A0P4VT57 A0A1B6DEN9 A0A1B6HYJ6 A0A1B6GH39 A0A146LWS2 A0A2H8TXB5 J9JWJ5 A0A067QTF9 A0A1B6KMY7 E0VIX9 A0A1W4WV82 A0A2H8TVN7 A0A2J7Q1Y7 A0A2J7Q1X1 A0A293MLS7 A0A293N5A1 A0A0P5FQT5 A0A0P5PJK7 A0A0P6G3C2 A0A0P4XNI7 A0A0N8CJ02 A0A2R5LMB4 A0A0N8A8B2 E9G2G2 A0A131YL43 L7M7U3 A0A131XFQ1 A0A1E1X4Y2 V5IBQ0 A0A1W4WJ56 A0A2P6KMS5 A0A224YMM6 A0A087US49 D6WW45 B7P4G4 V5HQ58 U4TYQ4 N6UH50 A0A232F0I2 A0A2P2I6K8 A0A0A1X0B2 A0A0K8VY37 A0A034V746 B5DPB0 B4LDS3 B3M5I6 A0A0N0U3D4 A0A2A3E819 A0A088A2Q3 W8BLS5 B4IZE4 A0A1W4UES8 A0A0P4WFK6 A0A1I8PUR4 B4KYE1 A0A1B0DNY5 A0A1V9XU82 A0A3B0KAW7 A0A3B0KG25 A0A3R7QVN1 A0A1B0CUG9 A0A1A9UG92 A0A0L7R7B6 A0A1B0FPI7 A0A336MQ60 A0A336MIU8 C3Y9B1 A0A0K8TLX6 A0A1I8JVJ1 A0A1Q3FD21 A0A182X4R0 A0A182KAK4 A0A182U1X3 A0A182I4Q9 Q5TQS7 A0A1S3HN93 A0A1S3IE79 B0X634 A0A182KNB5 A0A182H9F6 A0A182PHS1 A0A226DDK3 D5AEM7 A0A182IQX8 A0A0J9RSV2 H1UUN4 B3NFS2 Q9VSJ8 B4PG27 A0A1J1HJL0 A0A1L8DVA6 B4N6D7 B4HJI4
Pubmed
26354079
22118469
19121390
27129103
26823975
24845553
+ More
20566863 21292972 26830274 25576852 28049606 28503490 25765539 28797301 18362917 19820115 23537049 28648823 25830018 25348373 15632085 17994087 24495485 28327890 18563158 26369729 12364791 14747013 17210077 20966253 26483478 22936249 10731132 12537572 12537569 10908590 18327897 17550304
20566863 21292972 26830274 25576852 28049606 28503490 25765539 28797301 18362917 19820115 23537049 28648823 25830018 25348373 15632085 17994087 24495485 28327890 18563158 26369729 12364791 14747013 17210077 20966253 26483478 22936249 10731132 12537572 12537569 10908590 18327897 17550304
EMBL
KQ460205
KPJ17118.1
KQ459606
KPI91069.1
NWSH01005993
PCG63760.1
+ More
ODYU01012871 SOQ59385.1 AGBW02010220 OWR48897.1 BABH01036488 BABH01036489 BABH01036490 BABH01036491 BABH01036492 GDKW01000808 JAI55787.1 GEDC01013159 JAS24139.1 GECU01027986 JAS79720.1 GECZ01008010 JAS61759.1 GDHC01015744 GDHC01007829 JAQ02885.1 JAQ10800.1 GFXV01006764 MBW18569.1 ABLF02032299 KK852980 KDR13019.1 GEBQ01027177 JAT12800.1 DS235212 EEB13335.1 GFXV01006558 MBW18363.1 NEVH01019377 PNF22583.1 PNF22585.1 GFWV01017044 MAA41772.1 GFWV01023219 MAA47946.1 GDIQ01257569 JAJ94155.1 GDIQ01133077 JAL18649.1 GDIP01134678 GDIQ01039110 LRGB01000512 JAL69036.1 JAN55627.1 KZS18636.1 GDIP01242756 JAI80645.1 GDIP01127031 JAL76683.1 GGLE01006503 MBY10629.1 GDIP01172641 JAJ50761.1 GL732530 EFX86313.1 GEDV01008553 JAP80004.1 GACK01005092 JAA59942.1 GEFH01003623 JAP64958.1 GFAC01004900 JAT94288.1 GANP01015052 JAB69416.1 MWRG01008479 PRD27639.1 GFPF01007700 MAA18846.1 KK121308 KFM80188.1 KQ971361 EFA08190.1 ABJB010828408 DS635599 EEC01486.1 GANP01004259 GEFM01003850 JAB80209.1 JAP71946.1 KB631763 ERL85957.1 APGK01035808 KB740928 ENN77972.1 NNAY01001454 OXU23918.1 IACF01004067 LAB69663.1 GBXI01015216 GBXI01009538 JAC99075.1 JAD04754.1 GDHF01008476 JAI43838.1 GAKP01021589 JAC37363.1 CH379069 EDY73871.2 CH940647 EDW68946.2 CH902618 EDV39596.1 KQ435916 KOX68808.1 KZ288353 PBC27329.1 GAMC01008612 JAB97943.1 CH916366 EDV96699.1 GDRN01059976 JAI65451.1 CH933809 EDW17720.1 AJVK01007844 AJVK01007845 AJVK01007846 AJVK01007847 MNPL01004080 OQR76982.1 OUUW01000009 SPP85280.1 SPP85279.1 QCYY01001175 ROT79924.1 AJWK01029175 KQ414642 KOC66734.1 CCAG010022945 UFQS01002051 UFQT01002051 SSX13102.1 SSX32542.1 UFQT01001185 SSX29381.1 GG666492 EEN63167.1 GDAI01002244 JAI15359.1 GFDL01009561 JAV25484.1 APCN01002346 AAAB01008960 EAL40137.3 DS232400 EDS41165.1 JXUM01030164 KQ560874 KXJ80526.1 LNIX01000021 OXA43675.1 BT124764 ADF28805.1 CM002912 KMY98384.1 BT133119 AEY64287.1 CH954178 EDV50684.1 AE014296 AY069366 AAL39511.1 CM000159 EDW93171.1 CVRI01000006 CRK88213.1 GFDF01003710 JAV10374.1 CH964154 EDW79926.1 CH480815 EDW40708.1
ODYU01012871 SOQ59385.1 AGBW02010220 OWR48897.1 BABH01036488 BABH01036489 BABH01036490 BABH01036491 BABH01036492 GDKW01000808 JAI55787.1 GEDC01013159 JAS24139.1 GECU01027986 JAS79720.1 GECZ01008010 JAS61759.1 GDHC01015744 GDHC01007829 JAQ02885.1 JAQ10800.1 GFXV01006764 MBW18569.1 ABLF02032299 KK852980 KDR13019.1 GEBQ01027177 JAT12800.1 DS235212 EEB13335.1 GFXV01006558 MBW18363.1 NEVH01019377 PNF22583.1 PNF22585.1 GFWV01017044 MAA41772.1 GFWV01023219 MAA47946.1 GDIQ01257569 JAJ94155.1 GDIQ01133077 JAL18649.1 GDIP01134678 GDIQ01039110 LRGB01000512 JAL69036.1 JAN55627.1 KZS18636.1 GDIP01242756 JAI80645.1 GDIP01127031 JAL76683.1 GGLE01006503 MBY10629.1 GDIP01172641 JAJ50761.1 GL732530 EFX86313.1 GEDV01008553 JAP80004.1 GACK01005092 JAA59942.1 GEFH01003623 JAP64958.1 GFAC01004900 JAT94288.1 GANP01015052 JAB69416.1 MWRG01008479 PRD27639.1 GFPF01007700 MAA18846.1 KK121308 KFM80188.1 KQ971361 EFA08190.1 ABJB010828408 DS635599 EEC01486.1 GANP01004259 GEFM01003850 JAB80209.1 JAP71946.1 KB631763 ERL85957.1 APGK01035808 KB740928 ENN77972.1 NNAY01001454 OXU23918.1 IACF01004067 LAB69663.1 GBXI01015216 GBXI01009538 JAC99075.1 JAD04754.1 GDHF01008476 JAI43838.1 GAKP01021589 JAC37363.1 CH379069 EDY73871.2 CH940647 EDW68946.2 CH902618 EDV39596.1 KQ435916 KOX68808.1 KZ288353 PBC27329.1 GAMC01008612 JAB97943.1 CH916366 EDV96699.1 GDRN01059976 JAI65451.1 CH933809 EDW17720.1 AJVK01007844 AJVK01007845 AJVK01007846 AJVK01007847 MNPL01004080 OQR76982.1 OUUW01000009 SPP85280.1 SPP85279.1 QCYY01001175 ROT79924.1 AJWK01029175 KQ414642 KOC66734.1 CCAG010022945 UFQS01002051 UFQT01002051 SSX13102.1 SSX32542.1 UFQT01001185 SSX29381.1 GG666492 EEN63167.1 GDAI01002244 JAI15359.1 GFDL01009561 JAV25484.1 APCN01002346 AAAB01008960 EAL40137.3 DS232400 EDS41165.1 JXUM01030164 KQ560874 KXJ80526.1 LNIX01000021 OXA43675.1 BT124764 ADF28805.1 CM002912 KMY98384.1 BT133119 AEY64287.1 CH954178 EDV50684.1 AE014296 AY069366 AAL39511.1 CM000159 EDW93171.1 CVRI01000006 CRK88213.1 GFDF01003710 JAV10374.1 CH964154 EDW79926.1 CH480815 EDW40708.1
Proteomes
UP000053240
UP000053268
UP000218220
UP000007151
UP000079169
UP000005204
+ More
UP000007819 UP000027135 UP000009046 UP000192223 UP000235965 UP000076858 UP000000305 UP000054359 UP000007266 UP000001555 UP000030742 UP000019118 UP000215335 UP000001819 UP000008792 UP000007801 UP000053105 UP000242457 UP000005203 UP000001070 UP000192221 UP000095300 UP000009192 UP000092462 UP000192247 UP000268350 UP000283509 UP000092461 UP000078200 UP000053825 UP000092444 UP000001554 UP000075903 UP000076407 UP000075881 UP000075902 UP000075840 UP000007062 UP000085678 UP000002320 UP000075882 UP000069940 UP000249989 UP000075885 UP000198287 UP000075880 UP000008711 UP000000803 UP000002282 UP000183832 UP000007798 UP000001292
UP000007819 UP000027135 UP000009046 UP000192223 UP000235965 UP000076858 UP000000305 UP000054359 UP000007266 UP000001555 UP000030742 UP000019118 UP000215335 UP000001819 UP000008792 UP000007801 UP000053105 UP000242457 UP000005203 UP000001070 UP000192221 UP000095300 UP000009192 UP000092462 UP000192247 UP000268350 UP000283509 UP000092461 UP000078200 UP000053825 UP000092444 UP000001554 UP000075903 UP000076407 UP000075881 UP000075902 UP000075840 UP000007062 UP000085678 UP000002320 UP000075882 UP000069940 UP000249989 UP000075885 UP000198287 UP000075880 UP000008711 UP000000803 UP000002282 UP000183832 UP000007798 UP000001292
Interpro
Gene 3D
ProteinModelPortal
A0A194RH19
A0A194PEF4
A0A2A4IXA0
A0A2H1X227
A0A212F586
A0A3Q0JAX0
+ More
H9JVZ4 A0A0P4VT57 A0A1B6DEN9 A0A1B6HYJ6 A0A1B6GH39 A0A146LWS2 A0A2H8TXB5 J9JWJ5 A0A067QTF9 A0A1B6KMY7 E0VIX9 A0A1W4WV82 A0A2H8TVN7 A0A2J7Q1Y7 A0A2J7Q1X1 A0A293MLS7 A0A293N5A1 A0A0P5FQT5 A0A0P5PJK7 A0A0P6G3C2 A0A0P4XNI7 A0A0N8CJ02 A0A2R5LMB4 A0A0N8A8B2 E9G2G2 A0A131YL43 L7M7U3 A0A131XFQ1 A0A1E1X4Y2 V5IBQ0 A0A1W4WJ56 A0A2P6KMS5 A0A224YMM6 A0A087US49 D6WW45 B7P4G4 V5HQ58 U4TYQ4 N6UH50 A0A232F0I2 A0A2P2I6K8 A0A0A1X0B2 A0A0K8VY37 A0A034V746 B5DPB0 B4LDS3 B3M5I6 A0A0N0U3D4 A0A2A3E819 A0A088A2Q3 W8BLS5 B4IZE4 A0A1W4UES8 A0A0P4WFK6 A0A1I8PUR4 B4KYE1 A0A1B0DNY5 A0A1V9XU82 A0A3B0KAW7 A0A3B0KG25 A0A3R7QVN1 A0A1B0CUG9 A0A1A9UG92 A0A0L7R7B6 A0A1B0FPI7 A0A336MQ60 A0A336MIU8 C3Y9B1 A0A0K8TLX6 A0A1I8JVJ1 A0A1Q3FD21 A0A182X4R0 A0A182KAK4 A0A182U1X3 A0A182I4Q9 Q5TQS7 A0A1S3HN93 A0A1S3IE79 B0X634 A0A182KNB5 A0A182H9F6 A0A182PHS1 A0A226DDK3 D5AEM7 A0A182IQX8 A0A0J9RSV2 H1UUN4 B3NFS2 Q9VSJ8 B4PG27 A0A1J1HJL0 A0A1L8DVA6 B4N6D7 B4HJI4
H9JVZ4 A0A0P4VT57 A0A1B6DEN9 A0A1B6HYJ6 A0A1B6GH39 A0A146LWS2 A0A2H8TXB5 J9JWJ5 A0A067QTF9 A0A1B6KMY7 E0VIX9 A0A1W4WV82 A0A2H8TVN7 A0A2J7Q1Y7 A0A2J7Q1X1 A0A293MLS7 A0A293N5A1 A0A0P5FQT5 A0A0P5PJK7 A0A0P6G3C2 A0A0P4XNI7 A0A0N8CJ02 A0A2R5LMB4 A0A0N8A8B2 E9G2G2 A0A131YL43 L7M7U3 A0A131XFQ1 A0A1E1X4Y2 V5IBQ0 A0A1W4WJ56 A0A2P6KMS5 A0A224YMM6 A0A087US49 D6WW45 B7P4G4 V5HQ58 U4TYQ4 N6UH50 A0A232F0I2 A0A2P2I6K8 A0A0A1X0B2 A0A0K8VY37 A0A034V746 B5DPB0 B4LDS3 B3M5I6 A0A0N0U3D4 A0A2A3E819 A0A088A2Q3 W8BLS5 B4IZE4 A0A1W4UES8 A0A0P4WFK6 A0A1I8PUR4 B4KYE1 A0A1B0DNY5 A0A1V9XU82 A0A3B0KAW7 A0A3B0KG25 A0A3R7QVN1 A0A1B0CUG9 A0A1A9UG92 A0A0L7R7B6 A0A1B0FPI7 A0A336MQ60 A0A336MIU8 C3Y9B1 A0A0K8TLX6 A0A1I8JVJ1 A0A1Q3FD21 A0A182X4R0 A0A182KAK4 A0A182U1X3 A0A182I4Q9 Q5TQS7 A0A1S3HN93 A0A1S3IE79 B0X634 A0A182KNB5 A0A182H9F6 A0A182PHS1 A0A226DDK3 D5AEM7 A0A182IQX8 A0A0J9RSV2 H1UUN4 B3NFS2 Q9VSJ8 B4PG27 A0A1J1HJL0 A0A1L8DVA6 B4N6D7 B4HJI4
PDB
2PFT
E-value=4.56613e-10,
Score=148
Ontologies
GO
PANTHER
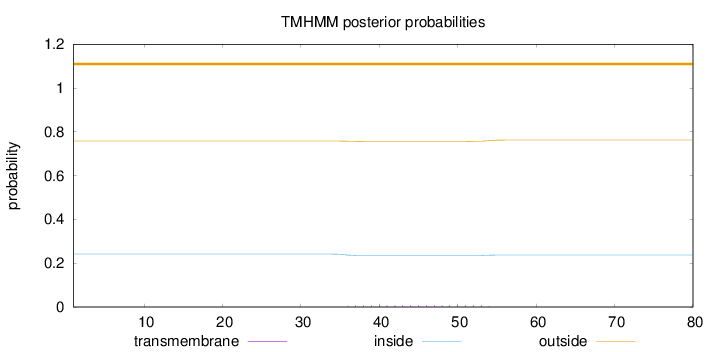
Topology
Length:
80
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.13712
Exp number, first 60 AAs:
0.13712
Total prob of N-in:
0.24169
outside
1 - 80
Population Genetic Test Statistics
Pi
261.371597
Theta
164.131645
Tajima's D
1.863235
CLR
0.237004
CSRT
0.859157042147893
Interpretation
Uncertain
