Gene
KWMTBOMO07593
Pre Gene Modal
BGIBMGA013714
Annotation
PREDICTED:_endothelin-converting_enzyme_1-like_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.171 Peroxisomal Reliability : 1.196
Sequence
CDS
ATGTATTACGGTTGCATCGAACGAACAGTAAGATGGTCTTCGGTGCTGATAGGAGCCTTAGGTGTGTCTCTGATCGCTCTCGCTATATACACCACAGTGCTGGTCCTCACGGATTACAAGAACCCGAAGCCCTGCTTGAGCGAGGCTTGTGTCAACACTGCGTCAAGAGTATTAGAAGCCCTCAATAAGGATGTGGATCCGTGCCACGACTTCTACGAGTTTGCCTGCGGAGGCTGGATCAAGAAGAACCCAGTACCAGAATGGGCTACATCTTGGGACCAGCTGGCGAAACTACGCGAACAACTAGTGGTAGATCTTCGAGAAGTTCTGGAGGCTGCAGACGACGAGGGTCTTCCACAGAGTGTGATCAAAGCGAAGAGCTTGTATAGAACTTGTATTGATACCGATAAATTGGAAGCTCAAGGGATTAAGCCCATCCAGGAACTTTTAGAAAAACTTGGCCTGCCTCCCACTCCACCAGTGAATGTCAGCGCCAACTTTACATGGGAGGATGTAGCTGGACGCGGAAGAAGGATGTTAGGCCTCAACGTGCTTCTCAGCATTCAGGTCGCTGAGGACGTGAGGAATACCAGCAGAAATCGTGTTGTGATCGAGCAAGTCCCTCCAGGCTTCAGCGAGCGCTACCTCCTCCGTCCGGAGCAGTTCAGCCACGAGATAGAACAGTACCATTTATACATTAAATCAGTGATCAGCATCGCCGACCCTGACACCGACGCCGAGAAGTTTGCTGATGACATTGTAGACTTCAGCAAGAGTCTAGCAAAGATAATGACCCCGGTGGAGGTGCGGCGCGGTGGCACTCATCTGTTTCACGAGGTTTCCGTGCGGCAGTTCGTGAAGGGTTCCGCCGGCGGACCCGCGCAGTGGGACCAGCACGACTGGGACAGGTATCTGGAGCTGGTGTTCGCCAACACGAGCGTGGGGCTGCAGCAGGACTCCGACCGAGTCATCGTGATGGACCTCCCGTACCTGCAGAAACTGGCCGTGCTGCTCAATGAGACCGAGCCTGTCATCGTCGAGCGCTTCGTGTGGTGGAGCGTATTCTCGACGGTGGCGCCCATGACGCTGAGCTCGTTCCGCACGCTGGGCTTCGAGTTCTCGCGCGCGGTGTTCGGGCTGCAGCAGCGCACGCCGCGCTGGAAGTCCTGCGCCTCCAACGTCAACGCCAACTTCGGCGTCGCGCTCAGCTACCTCTACGTCAAGAAACATTTCGACCAGGACTCCAGGCAGAAGGCCATAGAGATGATAGAGGACGTGCGCGAGGCCTTCTCCTCCAGCGTGCAGCGGCTGCGCTGGATGGACGCCGCCACGCGCGCGCGCACGCTGCGCAAGCTGGCCGCCATCAGGACCTTCGTGGGCTTCCCCTCCTGGCTGCTCACCCACCGGCAGCTCGACCAACATTACCAGCACGCGGAGGTGGTGGAGGGGAGCCTGTTCGGGACGTACCTGAAGCTGACCTGGGGCGCCGTGAAGAAATCTCTGGAGTCGCTCAGAGAGAAGCCCGACAGGAACAGGTGGGTGGCCACGGCGACCACTGTGAACGCGTTCTACTCGGCGACCCTCAACTCAGTCACGTTCCCCGCGGGCATCCTCCAGCCGCCCTTCTACGGGAACGGGATCGAAGCAATCAACTACGGTTCAATCGGCGCCATTATGGGTCACGAAGTGACGCATGGATTTGACGACCAAGGGCGTCGCTACGACGAGGAGGGCAACCTGAAGCAGTGGTGGTCGGCAGACACCCTCGAGCACTACCACGGCAAGGTGCAGTGCATCATCGATCAGTACAACAAGTACCACATGGCGCAGCTGCCCAACTACACGATAAACGGCCTGAACACGCAAGGCGAGAATATCGCCGACAACGGCGGAGTGCGCGCCGCCCTCGACGCCTACCGCCTGCACGCCGCCCGCGCGCCCGCCGCGTCTGCCGCGTCTGCCGCGCCCGCCGCCGACGCGCTGCCTGGACTGCCGCACTACTCGCCTACGCAGCTCTTCCACTTGGGATTCGCTCAGATCTGGTGCGGCAACTCCACCGTGGGAGCGCTCAAGTCTAAAATAGTAGAAGGCGTCCACAGCCCCAACAAGGTCCGAGTGATCGGCACCCTCAGCAACTCCGAGGAGTTTGCTGAAGCTTGGAAATGTCCCGTCGGGTCCCCCATGAACCCGGAGCACAAGTGTGTGCTCTGGTGA
Protein
MYYGCIERTVRWSSVLIGALGVSLIALAIYTTVLVLTDYKNPKPCLSEACVNTASRVLEALNKDVDPCHDFYEFACGGWIKKNPVPEWATSWDQLAKLREQLVVDLREVLEAADDEGLPQSVIKAKSLYRTCIDTDKLEAQGIKPIQELLEKLGLPPTPPVNVSANFTWEDVAGRGRRMLGLNVLLSIQVAEDVRNTSRNRVVIEQVPPGFSERYLLRPEQFSHEIEQYHLYIKSVISIADPDTDAEKFADDIVDFSKSLAKIMTPVEVRRGGTHLFHEVSVRQFVKGSAGGPAQWDQHDWDRYLELVFANTSVGLQQDSDRVIVMDLPYLQKLAVLLNETEPVIVERFVWWSVFSTVAPMTLSSFRTLGFEFSRAVFGLQQRTPRWKSCASNVNANFGVALSYLYVKKHFDQDSRQKAIEMIEDVREAFSSSVQRLRWMDAATRARTLRKLAAIRTFVGFPSWLLTHRQLDQHYQHAEVVEGSLFGTYLKLTWGAVKKSLESLREKPDRNRWVATATTVNAFYSATLNSVTFPAGILQPPFYGNGIEAINYGSIGAIMGHEVTHGFDDQGRRYDEEGNLKQWWSADTLEHYHGKVQCIIDQYNKYHMAQLPNYTINGLNTQGENIADNGGVRAALDAYRLHAARAPAASAASAAPAADALPGLPHYSPTQLFHLGFAQIWCGNSTVGALKSKIVEGVHSPNKVRVIGTLSNSEEFAEAWKCPVGSPMNPEHKCVLW
Summary
Uniprot
H9JW01
A0A2H1WLB0
A0A2A4JSI5
A0A3S2NQ70
A0A194PJ05
A0A212F708
+ More
S4PSE7 D6W7L0 A0A2J7R1H9 A0A2J7R1I2 A0A151HZV3 F4W4P9 A0A158NWD7 A0A151WUV5 E2BED8 E2ALK7 A0A195EVW1 A0A0L7RAM0 A0A195E2Y7 A0A0C9PKK5 A0A0C9QE15 A0A195CS15 A0A2A3EQM5 A0A088ANV7 A0A026W5J8 A0A3L8D3Q0 E9INR5 A0A2P8XRW8 A0A0N0BGE5 A0A0P5YC77 A0A0P5X1S1 A0A0P4ZW11 A0A0P4ZAW6 A0A2P8XEI8 A0A0P6IQY9 A0A0P5DKG3 A0A0P6BER9 A0A0P5SAE9 A0A3R7LPK0 A0A2R2MJ45 A0A2R2MJF6 A0A2R2MJ28 R7UA33 E9GJ71 E0VRJ2 A0A210PIH1 A0A164Q489 A0A162S6A1 A0A0P6C4X4 A0A0P6DRD0 A0A164K4K6 A0A336KGD2 A0A0P6IWU2 A0A088AKD7 A0A0P5VRQ9 E9H0B2 E9H6H7 A0A0P4ZXV9 Q17AS1 A0A0P6FPK3 B0W3N5 A0A3B4ZX63 A0A0P6AK73 A0A0N8AD00 A0A0P6DAF3 A0A0P6FL55 A0A0N8A4R4 A0A2U8JG00 A0A0P6FGR2 A0A0P5JFY3 A0A194QVE0 A0A0L7R038 A0A016WY34 S4NTA7 A0A3B3SIA4 A0A3Q1B1Z4 A0A0P5KX79 A0A0P5LDE2 A0A0P5NQC1 A0A3P8TQJ8 A0A3B3SH04 A0A0P5T3J5 A0A0N8BA02 A0A3P8TMN2 A0A0P6FJS1 A0A3B3SI52 T1I287 A0A0P5M453 K7J3Q5 A0A2R5LIL2 A0A0P6FKW5 A0A0P5UVQ0 A0A0P5MW22 A0A0P5V268 A0A0K8S8J1
S4PSE7 D6W7L0 A0A2J7R1H9 A0A2J7R1I2 A0A151HZV3 F4W4P9 A0A158NWD7 A0A151WUV5 E2BED8 E2ALK7 A0A195EVW1 A0A0L7RAM0 A0A195E2Y7 A0A0C9PKK5 A0A0C9QE15 A0A195CS15 A0A2A3EQM5 A0A088ANV7 A0A026W5J8 A0A3L8D3Q0 E9INR5 A0A2P8XRW8 A0A0N0BGE5 A0A0P5YC77 A0A0P5X1S1 A0A0P4ZW11 A0A0P4ZAW6 A0A2P8XEI8 A0A0P6IQY9 A0A0P5DKG3 A0A0P6BER9 A0A0P5SAE9 A0A3R7LPK0 A0A2R2MJ45 A0A2R2MJF6 A0A2R2MJ28 R7UA33 E9GJ71 E0VRJ2 A0A210PIH1 A0A164Q489 A0A162S6A1 A0A0P6C4X4 A0A0P6DRD0 A0A164K4K6 A0A336KGD2 A0A0P6IWU2 A0A088AKD7 A0A0P5VRQ9 E9H0B2 E9H6H7 A0A0P4ZXV9 Q17AS1 A0A0P6FPK3 B0W3N5 A0A3B4ZX63 A0A0P6AK73 A0A0N8AD00 A0A0P6DAF3 A0A0P6FL55 A0A0N8A4R4 A0A2U8JG00 A0A0P6FGR2 A0A0P5JFY3 A0A194QVE0 A0A0L7R038 A0A016WY34 S4NTA7 A0A3B3SIA4 A0A3Q1B1Z4 A0A0P5KX79 A0A0P5LDE2 A0A0P5NQC1 A0A3P8TQJ8 A0A3B3SH04 A0A0P5T3J5 A0A0N8BA02 A0A3P8TMN2 A0A0P6FJS1 A0A3B3SI52 T1I287 A0A0P5M453 K7J3Q5 A0A2R5LIL2 A0A0P6FKW5 A0A0P5UVQ0 A0A0P5MW22 A0A0P5V268 A0A0K8S8J1
Pubmed
EMBL
BABH01036499
BABH01036500
BABH01036501
ODYU01009064
SOQ53214.1
NWSH01000725
+ More
PCG74574.1 RSAL01000433 RVE41737.1 KQ459606 KPI91065.1 AGBW02009952 OWR49508.1 GAIX01013918 JAA78642.1 KQ971307 EFA11308.2 NEVH01008206 PNF34683.1 PNF34684.1 KQ976657 KYM78626.1 GL887547 EGI70815.1 ADTU01027908 ADTU01027909 KQ982718 KYQ51710.1 GL447768 EFN85983.1 GL440609 EFN65709.1 KQ981954 KYN32037.1 KQ414618 KOC67962.1 KQ979701 KYN19530.1 GBYB01001558 JAG71325.1 GBYB01001559 GBYB01001561 JAG71326.1 JAG71328.1 KQ977329 KYN03491.1 KZ288193 PBC34065.1 KK107405 EZA51278.1 QOIP01000014 RLU14856.1 GL764397 EFZ17786.1 PYGN01001454 PSN34745.1 KQ435783 KOX74681.1 GDIP01147629 GDIP01060541 GDIP01038997 JAM43174.1 GDIP01078841 JAM24874.1 GDIP01207819 JAJ15583.1 GDIP01215122 JAJ08280.1 PYGN01002487 PSN30408.1 GDIQ01001382 JAN93355.1 GDIP01158285 JAJ65117.1 GDIP01015664 JAM88051.1 GDIQ01098833 JAL52893.1 QCYY01004450 ROT60976.1 AMQN01009837 KB306314 ELT99995.1 GL732547 EFX80396.1 DS235478 EEB15998.1 NEDP02076649 OWF36277.1 LRGB01002481 KZS07419.1 LRGB01000084 KZS21047.1 GDIP01005817 JAM97898.1 GDIQ01074176 JAN20561.1 LRGB01003375 KZS02943.1 UFQS01000279 UFQS01001601 UFQT01000279 UFQT01001601 SSX02335.1 SSX11555.1 SSX22710.1 GDIQ01026654 JAN68083.1 GDIP01110774 JAL92940.1 GL732580 EFX74773.1 GL732597 EFX72698.1 GDIP01206573 JAJ16829.1 CH477330 EAT43387.1 GDIQ01047301 JAN47436.1 DS231833 EDS32182.1 GDIP01028145 JAM75570.1 GDIP01159537 JAJ63865.1 GDIQ01081042 JAN13695.1 GDIQ01048800 JAN45937.1 GDIP01182625 JAJ40777.1 MH298329 AWK67618.1 GDIQ01048801 JAN45936.1 GDIQ01200196 JAK51529.1 KQ461108 KPJ09289.1 KQ414672 KOC64187.1 JARK01000051 EYC44744.1 GAIX01012266 JAA80294.1 GDIQ01178792 GDIQ01111970 JAK72933.1 GDIQ01178791 JAK72934.1 GDIQ01180249 GDIQ01145877 JAL05849.1 GDIQ01195099 GDIQ01096930 GDIQ01046417 JAL54796.1 GDIQ01234233 GDIQ01198243 JAK53482.1 GDIQ01213584 GDIQ01096931 GDIQ01063401 GDIQ01046416 JAN48321.1 ACPB03014798 GDIQ01161117 GDIQ01080393 GDIQ01080392 JAK90608.1 GGLE01005244 MBY09370.1 GDIQ01240630 GDIQ01113374 GDIQ01048010 JAN46727.1 GDIP01108036 JAL95678.1 GDIQ01152710 GDIQ01136973 JAK99015.1 GDIP01105531 JAL98183.1 GBRD01016233 JAG49593.1
PCG74574.1 RSAL01000433 RVE41737.1 KQ459606 KPI91065.1 AGBW02009952 OWR49508.1 GAIX01013918 JAA78642.1 KQ971307 EFA11308.2 NEVH01008206 PNF34683.1 PNF34684.1 KQ976657 KYM78626.1 GL887547 EGI70815.1 ADTU01027908 ADTU01027909 KQ982718 KYQ51710.1 GL447768 EFN85983.1 GL440609 EFN65709.1 KQ981954 KYN32037.1 KQ414618 KOC67962.1 KQ979701 KYN19530.1 GBYB01001558 JAG71325.1 GBYB01001559 GBYB01001561 JAG71326.1 JAG71328.1 KQ977329 KYN03491.1 KZ288193 PBC34065.1 KK107405 EZA51278.1 QOIP01000014 RLU14856.1 GL764397 EFZ17786.1 PYGN01001454 PSN34745.1 KQ435783 KOX74681.1 GDIP01147629 GDIP01060541 GDIP01038997 JAM43174.1 GDIP01078841 JAM24874.1 GDIP01207819 JAJ15583.1 GDIP01215122 JAJ08280.1 PYGN01002487 PSN30408.1 GDIQ01001382 JAN93355.1 GDIP01158285 JAJ65117.1 GDIP01015664 JAM88051.1 GDIQ01098833 JAL52893.1 QCYY01004450 ROT60976.1 AMQN01009837 KB306314 ELT99995.1 GL732547 EFX80396.1 DS235478 EEB15998.1 NEDP02076649 OWF36277.1 LRGB01002481 KZS07419.1 LRGB01000084 KZS21047.1 GDIP01005817 JAM97898.1 GDIQ01074176 JAN20561.1 LRGB01003375 KZS02943.1 UFQS01000279 UFQS01001601 UFQT01000279 UFQT01001601 SSX02335.1 SSX11555.1 SSX22710.1 GDIQ01026654 JAN68083.1 GDIP01110774 JAL92940.1 GL732580 EFX74773.1 GL732597 EFX72698.1 GDIP01206573 JAJ16829.1 CH477330 EAT43387.1 GDIQ01047301 JAN47436.1 DS231833 EDS32182.1 GDIP01028145 JAM75570.1 GDIP01159537 JAJ63865.1 GDIQ01081042 JAN13695.1 GDIQ01048800 JAN45937.1 GDIP01182625 JAJ40777.1 MH298329 AWK67618.1 GDIQ01048801 JAN45936.1 GDIQ01200196 JAK51529.1 KQ461108 KPJ09289.1 KQ414672 KOC64187.1 JARK01000051 EYC44744.1 GAIX01012266 JAA80294.1 GDIQ01178792 GDIQ01111970 JAK72933.1 GDIQ01178791 JAK72934.1 GDIQ01180249 GDIQ01145877 JAL05849.1 GDIQ01195099 GDIQ01096930 GDIQ01046417 JAL54796.1 GDIQ01234233 GDIQ01198243 JAK53482.1 GDIQ01213584 GDIQ01096931 GDIQ01063401 GDIQ01046416 JAN48321.1 ACPB03014798 GDIQ01161117 GDIQ01080393 GDIQ01080392 JAK90608.1 GGLE01005244 MBY09370.1 GDIQ01240630 GDIQ01113374 GDIQ01048010 JAN46727.1 GDIP01108036 JAL95678.1 GDIQ01152710 GDIQ01136973 JAK99015.1 GDIP01105531 JAL98183.1 GBRD01016233 JAG49593.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000007151
UP000007266
+ More
UP000235965 UP000078540 UP000007755 UP000005205 UP000075809 UP000008237 UP000000311 UP000078541 UP000053825 UP000078492 UP000078542 UP000242457 UP000005203 UP000053097 UP000279307 UP000245037 UP000053105 UP000283509 UP000085678 UP000014760 UP000000305 UP000009046 UP000242188 UP000076858 UP000008820 UP000002320 UP000261400 UP000053240 UP000024635 UP000261540 UP000257160 UP000265080 UP000015103 UP000002358
UP000235965 UP000078540 UP000007755 UP000005205 UP000075809 UP000008237 UP000000311 UP000078541 UP000053825 UP000078492 UP000078542 UP000242457 UP000005203 UP000053097 UP000279307 UP000245037 UP000053105 UP000283509 UP000085678 UP000014760 UP000000305 UP000009046 UP000242188 UP000076858 UP000008820 UP000002320 UP000261400 UP000053240 UP000024635 UP000261540 UP000257160 UP000265080 UP000015103 UP000002358
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JW01
A0A2H1WLB0
A0A2A4JSI5
A0A3S2NQ70
A0A194PJ05
A0A212F708
+ More
S4PSE7 D6W7L0 A0A2J7R1H9 A0A2J7R1I2 A0A151HZV3 F4W4P9 A0A158NWD7 A0A151WUV5 E2BED8 E2ALK7 A0A195EVW1 A0A0L7RAM0 A0A195E2Y7 A0A0C9PKK5 A0A0C9QE15 A0A195CS15 A0A2A3EQM5 A0A088ANV7 A0A026W5J8 A0A3L8D3Q0 E9INR5 A0A2P8XRW8 A0A0N0BGE5 A0A0P5YC77 A0A0P5X1S1 A0A0P4ZW11 A0A0P4ZAW6 A0A2P8XEI8 A0A0P6IQY9 A0A0P5DKG3 A0A0P6BER9 A0A0P5SAE9 A0A3R7LPK0 A0A2R2MJ45 A0A2R2MJF6 A0A2R2MJ28 R7UA33 E9GJ71 E0VRJ2 A0A210PIH1 A0A164Q489 A0A162S6A1 A0A0P6C4X4 A0A0P6DRD0 A0A164K4K6 A0A336KGD2 A0A0P6IWU2 A0A088AKD7 A0A0P5VRQ9 E9H0B2 E9H6H7 A0A0P4ZXV9 Q17AS1 A0A0P6FPK3 B0W3N5 A0A3B4ZX63 A0A0P6AK73 A0A0N8AD00 A0A0P6DAF3 A0A0P6FL55 A0A0N8A4R4 A0A2U8JG00 A0A0P6FGR2 A0A0P5JFY3 A0A194QVE0 A0A0L7R038 A0A016WY34 S4NTA7 A0A3B3SIA4 A0A3Q1B1Z4 A0A0P5KX79 A0A0P5LDE2 A0A0P5NQC1 A0A3P8TQJ8 A0A3B3SH04 A0A0P5T3J5 A0A0N8BA02 A0A3P8TMN2 A0A0P6FJS1 A0A3B3SI52 T1I287 A0A0P5M453 K7J3Q5 A0A2R5LIL2 A0A0P6FKW5 A0A0P5UVQ0 A0A0P5MW22 A0A0P5V268 A0A0K8S8J1
S4PSE7 D6W7L0 A0A2J7R1H9 A0A2J7R1I2 A0A151HZV3 F4W4P9 A0A158NWD7 A0A151WUV5 E2BED8 E2ALK7 A0A195EVW1 A0A0L7RAM0 A0A195E2Y7 A0A0C9PKK5 A0A0C9QE15 A0A195CS15 A0A2A3EQM5 A0A088ANV7 A0A026W5J8 A0A3L8D3Q0 E9INR5 A0A2P8XRW8 A0A0N0BGE5 A0A0P5YC77 A0A0P5X1S1 A0A0P4ZW11 A0A0P4ZAW6 A0A2P8XEI8 A0A0P6IQY9 A0A0P5DKG3 A0A0P6BER9 A0A0P5SAE9 A0A3R7LPK0 A0A2R2MJ45 A0A2R2MJF6 A0A2R2MJ28 R7UA33 E9GJ71 E0VRJ2 A0A210PIH1 A0A164Q489 A0A162S6A1 A0A0P6C4X4 A0A0P6DRD0 A0A164K4K6 A0A336KGD2 A0A0P6IWU2 A0A088AKD7 A0A0P5VRQ9 E9H0B2 E9H6H7 A0A0P4ZXV9 Q17AS1 A0A0P6FPK3 B0W3N5 A0A3B4ZX63 A0A0P6AK73 A0A0N8AD00 A0A0P6DAF3 A0A0P6FL55 A0A0N8A4R4 A0A2U8JG00 A0A0P6FGR2 A0A0P5JFY3 A0A194QVE0 A0A0L7R038 A0A016WY34 S4NTA7 A0A3B3SIA4 A0A3Q1B1Z4 A0A0P5KX79 A0A0P5LDE2 A0A0P5NQC1 A0A3P8TQJ8 A0A3B3SH04 A0A0P5T3J5 A0A0N8BA02 A0A3P8TMN2 A0A0P6FJS1 A0A3B3SI52 T1I287 A0A0P5M453 K7J3Q5 A0A2R5LIL2 A0A0P6FKW5 A0A0P5UVQ0 A0A0P5MW22 A0A0P5V268 A0A0K8S8J1
PDB
3DWB
E-value=8.63874e-113,
Score=1043
Ontologies
GO
PANTHER
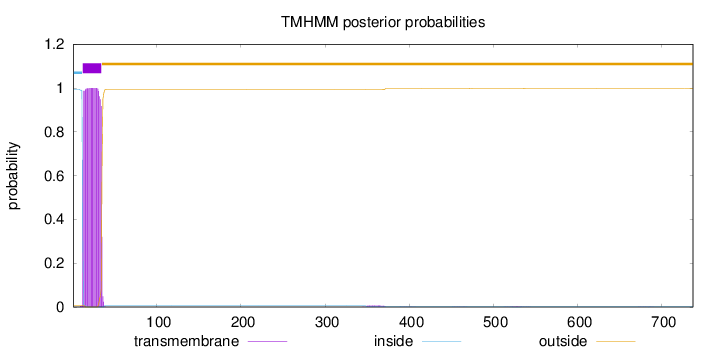
Topology
Length:
737
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.04117
Exp number, first 60 AAs:
22.85277
Total prob of N-in:
0.99425
POSSIBLE N-term signal
sequence
inside
1 - 11
TMhelix
12 - 34
outside
35 - 737
Population Genetic Test Statistics
Pi
312.132641
Theta
223.867365
Tajima's D
0.504618
CLR
0.463161
CSRT
0.515074246287686
Interpretation
Uncertain
