Gene
KWMTBOMO07592
Pre Gene Modal
BGIBMGA013703
Annotation
UTP11-like_U3_small_nucleolar_ribonucleoprotein_[Bombyx_mori]
Full name
U3 small nucleolar RNA-associated protein 11
Location in the cell
Nuclear Reliability : 4.316
Sequence
CDS
ATGTCGTCGTGGAAGAAGGCCGCTAAGGCGAATCAGAAAACTCACAGAGAACGACATCAACCAGAAGCTCGGAAACATCTTGGACTTCTCGAAAAGAAGAAGGATTATAAAAAGAGAGCAGATGATTACCATGAGAAAGGTCAAACCTTAAAACTTCTTCGGAAAAGGACACTTGATAAGAACCCAGATGAATTTTACTATCACATGATAAACTCTAGGGTGAAAGAAGGGGTACATCATGAGCTCCAGAAAGAGGGTGAGCACTCGTCGGAGCAGGTGAAGTTGATGCAGACTCAAGATATCAAATATATCAACATGAAACGCACCATTGAAAGTAGAAGAATCAATAGACTACAGTCCCAACTTCATATGACAGACGTAGCAGAGGCGACACCAAACACACACACATTCTTTGTTGATGAAGGTGAAGAGAATGGCTTTGATTTAGCGAAGAGACTTGACACACATCCTGCTCTGATTGGCAGAAAATCCAATAGGCCAAGATTGTCTGACTTAAACAAGATAACTCTACCAGACTTGGATGAAGAGACTGTAAAAGAAATGAAAAAGAAAAAGGAGAAAGTTTACAAAGAGATTGCGAAAAGAATTCAAAGAGAGACCGAGTTGACAGTTGTCCAACAAAAGATGGAGCTAAAACGTCACCTGCAGGACACTTCAGTACTAAAGCCAAAGAAAATAAAGAAGGGTTCGGCAAAAGCGGCTCCAGTTTACAAATTCCATTACATAAGGAAGAAATAA
Protein
MSSWKKAAKANQKTHRERHQPEARKHLGLLEKKKDYKKRADDYHEKGQTLKLLRKRTLDKNPDEFYYHMINSRVKEGVHHELQKEGEHSSEQVKLMQTQDIKYINMKRTIESRRINRLQSQLHMTDVAEATPNTHTFFVDEGEENGFDLAKRLDTHPALIGRKSNRPRLSDLNKITLPDLDEETVKEMKKKKEKVYKEIAKRIQRETELTVVQQKMELKRHLQDTSVLKPKKIKKGSAKAAPVYKFHYIRKK
Summary
Description
Involved in nucleolar processing of pre-18S ribosomal RNA.
Subunit
Component of the ribosomal small subunit (SSU) processome.
Similarity
Belongs to the UTP11 family.
Uniprot
Q1HPT7
A0A2H1WJF8
A0A1E1VZ43
A0A3S2TC82
A0A2A4JS36
A0A194RI46
+ More
A0A194PEE9 S4P852 A0A212F720 A0A154P8E9 Q16VF9 A0A195F3D7 A0A1A9VRF1 A0A182S705 A0A182HE04 D3TQW5 A0A151IXT0 A0A2M4BXD9 A0A0L7R232 A0A2P8XXK5 A0A182W325 A0A195BDE7 A0A158NZR6 W5JH54 A0A182IJ22 A0A182YJN2 A0A2M3ZJF1 A0A182MMJ8 A0A151WY04 A0A2M4AXL7 A0A182KEH7 A0A084VFV0 A0A336LL99 A0A1A9WX10 A0A182P3G7 F4WLB6 A0A023EL89 A0A182F772 E2BPM4 A0A088AE86 A0A0L7KN25 A0A1A9XDV8 A0A182R8F6 A0A1B0B8H6 A0A0C9RBL6 A0A1J1HFN6 A0A1I8M9S7 A0A0M9A129 E2A6K8 A0A182UTI9 A0A182LNH4 A0A182HVH7 A0A182XF10 A0A0K8VRH6 A0A232F9F0 A0A026W318 K7IP59 Q7PQ85 A0A034VRH1 A0A067RCZ4 A0A182TT63 A0A1B0G0S1 W8B522 E0VN76 A0A2J7PEY7 A0A1I8PCR2 A0A182QRQ8 A0A182NCN7 A0A0A1XI80 A0A0J7N5N7 A0A1Q3F1Z5 B0W3D5 C4WYC0 A0A2A3ETA8 A0A195D1Q6 A0A0L0CQH3 A0A1B6LFV5 A3EXT0 A0A1B6EZH7 A0A0P6H4Q3 A0A1Z5KUS8 A0A1B6I2Q6 A0A0P5HKF2 A0A0P5XC94 A0A0P6B709 E9H9I9 A0A2R5LIA9 A0A0P6JPZ3 A0A0P5ZJB1 A0A293LJ02 A0A131YBB6 A0A0K8RL62 A0A0P5IS99 B7Q8Y0 A0A1B6DD35 L7M7F2 A0A1S3I255 A0A224Z2Q1 A0A0C9R1S0
A0A194PEE9 S4P852 A0A212F720 A0A154P8E9 Q16VF9 A0A195F3D7 A0A1A9VRF1 A0A182S705 A0A182HE04 D3TQW5 A0A151IXT0 A0A2M4BXD9 A0A0L7R232 A0A2P8XXK5 A0A182W325 A0A195BDE7 A0A158NZR6 W5JH54 A0A182IJ22 A0A182YJN2 A0A2M3ZJF1 A0A182MMJ8 A0A151WY04 A0A2M4AXL7 A0A182KEH7 A0A084VFV0 A0A336LL99 A0A1A9WX10 A0A182P3G7 F4WLB6 A0A023EL89 A0A182F772 E2BPM4 A0A088AE86 A0A0L7KN25 A0A1A9XDV8 A0A182R8F6 A0A1B0B8H6 A0A0C9RBL6 A0A1J1HFN6 A0A1I8M9S7 A0A0M9A129 E2A6K8 A0A182UTI9 A0A182LNH4 A0A182HVH7 A0A182XF10 A0A0K8VRH6 A0A232F9F0 A0A026W318 K7IP59 Q7PQ85 A0A034VRH1 A0A067RCZ4 A0A182TT63 A0A1B0G0S1 W8B522 E0VN76 A0A2J7PEY7 A0A1I8PCR2 A0A182QRQ8 A0A182NCN7 A0A0A1XI80 A0A0J7N5N7 A0A1Q3F1Z5 B0W3D5 C4WYC0 A0A2A3ETA8 A0A195D1Q6 A0A0L0CQH3 A0A1B6LFV5 A3EXT0 A0A1B6EZH7 A0A0P6H4Q3 A0A1Z5KUS8 A0A1B6I2Q6 A0A0P5HKF2 A0A0P5XC94 A0A0P6B709 E9H9I9 A0A2R5LIA9 A0A0P6JPZ3 A0A0P5ZJB1 A0A293LJ02 A0A131YBB6 A0A0K8RL62 A0A0P5IS99 B7Q8Y0 A0A1B6DD35 L7M7F2 A0A1S3I255 A0A224Z2Q1 A0A0C9R1S0
Pubmed
19121390
26354079
23622113
22118469
17510324
26483478
+ More
20353571 29403074 21347285 20920257 23761445 25244985 24438588 21719571 24945155 20798317 26227816 25315136 20966253 28648823 24508170 30249741 20075255 12364791 14747013 17210077 25348373 24845553 24495485 20566863 25830018 26108605 28528879 21292972 25576852 28797301 26131772
20353571 29403074 21347285 20920257 23761445 25244985 24438588 21719571 24945155 20798317 26227816 25315136 20966253 28648823 24508170 30249741 20075255 12364791 14747013 17210077 25348373 24845553 24495485 20566863 25830018 26108605 28528879 21292972 25576852 28797301 26131772
EMBL
BABH01036501
BABH01036502
DQ443315
ABF51404.1
ODYU01009064
SOQ53213.1
+ More
GDQN01011054 JAT80000.1 RSAL01000433 RVE41738.1 NWSH01000725 PCG74576.1 KQ460205 KPJ17109.1 KQ459606 KPI91064.1 GAIX01006096 JAA86464.1 AGBW02009952 OWR49509.1 KQ434827 KZC07488.1 CH477594 EAT38544.1 KQ981856 KYN34692.1 JXUM01034813 KQ561037 KXJ79935.1 EZ423817 ADD20093.1 KQ980791 KYN13123.1 GGFJ01008578 MBW57719.1 KQ414666 KOC64899.1 PYGN01001197 PSN36738.1 KQ976511 KYM82583.1 ADTU01004931 ADMH02001534 ETN62144.1 GGFM01007881 MBW28632.1 AXCM01000316 KQ982656 KYQ52743.1 GGFK01012131 MBW45452.1 ATLV01012488 KE524796 KFB36844.1 UFQT01000042 SSX18736.1 GL888207 EGI65079.1 GAPW01003622 JAC09976.1 GL449633 EFN82428.1 JTDY01008612 KOB64486.1 JXJN01009945 GBYB01013919 JAG83686.1 CVRI01000002 CRK86783.1 KQ435794 KOX73649.1 GL437134 EFN70923.1 APCN01005778 GDHF01011144 JAI41170.1 NNAY01000633 OXU27295.1 KK107488 QOIP01000012 EZA49981.1 RLU15651.1 AAAB01008898 EAA09174.5 GAKP01013047 GAKP01013046 JAC45906.1 KK852547 KDR21607.1 CCAG010004515 GAMC01021666 JAB84889.1 DS235335 EEB14832.1 NEVH01026086 PNF14900.1 AXCN02001182 GBXI01003625 JAD10667.1 LBMM01009588 KMQ88010.1 GFDL01013469 JAV21576.1 DS231831 EDS31278.1 ABLF02009719 AK343117 BAH72890.1 KZ288192 PBC34379.1 KQ976956 KYN06838.1 JRES01000062 KNC34496.1 GEBQ01017418 JAT22559.1 EF091998 ABN11990.1 GECZ01026548 JAS43221.1 GDIQ01025266 JAN69471.1 GFJQ02008326 JAV98643.1 GECU01026561 JAS81145.1 GDIQ01231814 JAK19911.1 GDIP01074220 JAM29495.1 GDIP01021078 JAM82637.1 GL732608 EFX71625.1 GGLE01004931 MBY09057.1 GDIQ01012656 JAN82081.1 GDIP01042819 JAM60896.1 GFWV01003306 MAA28036.1 GEFM01000420 JAP75376.1 GADI01002185 JAA71623.1 GDIQ01209775 JAK41950.1 ABJB010094388 ABJB010486046 DS886620 EEC15302.1 GEDC01013694 JAS23604.1 GACK01005981 JAA59053.1 GFPF01012990 MAA24136.1 GBZX01001765 JAG90975.1
GDQN01011054 JAT80000.1 RSAL01000433 RVE41738.1 NWSH01000725 PCG74576.1 KQ460205 KPJ17109.1 KQ459606 KPI91064.1 GAIX01006096 JAA86464.1 AGBW02009952 OWR49509.1 KQ434827 KZC07488.1 CH477594 EAT38544.1 KQ981856 KYN34692.1 JXUM01034813 KQ561037 KXJ79935.1 EZ423817 ADD20093.1 KQ980791 KYN13123.1 GGFJ01008578 MBW57719.1 KQ414666 KOC64899.1 PYGN01001197 PSN36738.1 KQ976511 KYM82583.1 ADTU01004931 ADMH02001534 ETN62144.1 GGFM01007881 MBW28632.1 AXCM01000316 KQ982656 KYQ52743.1 GGFK01012131 MBW45452.1 ATLV01012488 KE524796 KFB36844.1 UFQT01000042 SSX18736.1 GL888207 EGI65079.1 GAPW01003622 JAC09976.1 GL449633 EFN82428.1 JTDY01008612 KOB64486.1 JXJN01009945 GBYB01013919 JAG83686.1 CVRI01000002 CRK86783.1 KQ435794 KOX73649.1 GL437134 EFN70923.1 APCN01005778 GDHF01011144 JAI41170.1 NNAY01000633 OXU27295.1 KK107488 QOIP01000012 EZA49981.1 RLU15651.1 AAAB01008898 EAA09174.5 GAKP01013047 GAKP01013046 JAC45906.1 KK852547 KDR21607.1 CCAG010004515 GAMC01021666 JAB84889.1 DS235335 EEB14832.1 NEVH01026086 PNF14900.1 AXCN02001182 GBXI01003625 JAD10667.1 LBMM01009588 KMQ88010.1 GFDL01013469 JAV21576.1 DS231831 EDS31278.1 ABLF02009719 AK343117 BAH72890.1 KZ288192 PBC34379.1 KQ976956 KYN06838.1 JRES01000062 KNC34496.1 GEBQ01017418 JAT22559.1 EF091998 ABN11990.1 GECZ01026548 JAS43221.1 GDIQ01025266 JAN69471.1 GFJQ02008326 JAV98643.1 GECU01026561 JAS81145.1 GDIQ01231814 JAK19911.1 GDIP01074220 JAM29495.1 GDIP01021078 JAM82637.1 GL732608 EFX71625.1 GGLE01004931 MBY09057.1 GDIQ01012656 JAN82081.1 GDIP01042819 JAM60896.1 GFWV01003306 MAA28036.1 GEFM01000420 JAP75376.1 GADI01002185 JAA71623.1 GDIQ01209775 JAK41950.1 ABJB010094388 ABJB010486046 DS886620 EEC15302.1 GEDC01013694 JAS23604.1 GACK01005981 JAA59053.1 GFPF01012990 MAA24136.1 GBZX01001765 JAG90975.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053240
UP000053268
UP000007151
+ More
UP000076502 UP000008820 UP000078541 UP000078200 UP000075901 UP000069940 UP000249989 UP000078492 UP000053825 UP000245037 UP000075920 UP000078540 UP000005205 UP000000673 UP000075880 UP000076408 UP000075883 UP000075809 UP000075881 UP000030765 UP000091820 UP000075885 UP000007755 UP000069272 UP000008237 UP000005203 UP000037510 UP000092443 UP000075900 UP000092460 UP000183832 UP000095301 UP000053105 UP000000311 UP000075903 UP000075882 UP000075840 UP000076407 UP000215335 UP000053097 UP000279307 UP000002358 UP000007062 UP000027135 UP000075902 UP000092444 UP000009046 UP000235965 UP000095300 UP000075886 UP000075884 UP000036403 UP000002320 UP000007819 UP000242457 UP000078542 UP000037069 UP000000305 UP000001555 UP000085678
UP000076502 UP000008820 UP000078541 UP000078200 UP000075901 UP000069940 UP000249989 UP000078492 UP000053825 UP000245037 UP000075920 UP000078540 UP000005205 UP000000673 UP000075880 UP000076408 UP000075883 UP000075809 UP000075881 UP000030765 UP000091820 UP000075885 UP000007755 UP000069272 UP000008237 UP000005203 UP000037510 UP000092443 UP000075900 UP000092460 UP000183832 UP000095301 UP000053105 UP000000311 UP000075903 UP000075882 UP000075840 UP000076407 UP000215335 UP000053097 UP000279307 UP000002358 UP000007062 UP000027135 UP000075902 UP000092444 UP000009046 UP000235965 UP000095300 UP000075886 UP000075884 UP000036403 UP000002320 UP000007819 UP000242457 UP000078542 UP000037069 UP000000305 UP000001555 UP000085678
PRIDE
Pfam
PF03998 Utp11
Interpro
IPR007144
SSU_processome_Utp11
ProteinModelPortal
Q1HPT7
A0A2H1WJF8
A0A1E1VZ43
A0A3S2TC82
A0A2A4JS36
A0A194RI46
+ More
A0A194PEE9 S4P852 A0A212F720 A0A154P8E9 Q16VF9 A0A195F3D7 A0A1A9VRF1 A0A182S705 A0A182HE04 D3TQW5 A0A151IXT0 A0A2M4BXD9 A0A0L7R232 A0A2P8XXK5 A0A182W325 A0A195BDE7 A0A158NZR6 W5JH54 A0A182IJ22 A0A182YJN2 A0A2M3ZJF1 A0A182MMJ8 A0A151WY04 A0A2M4AXL7 A0A182KEH7 A0A084VFV0 A0A336LL99 A0A1A9WX10 A0A182P3G7 F4WLB6 A0A023EL89 A0A182F772 E2BPM4 A0A088AE86 A0A0L7KN25 A0A1A9XDV8 A0A182R8F6 A0A1B0B8H6 A0A0C9RBL6 A0A1J1HFN6 A0A1I8M9S7 A0A0M9A129 E2A6K8 A0A182UTI9 A0A182LNH4 A0A182HVH7 A0A182XF10 A0A0K8VRH6 A0A232F9F0 A0A026W318 K7IP59 Q7PQ85 A0A034VRH1 A0A067RCZ4 A0A182TT63 A0A1B0G0S1 W8B522 E0VN76 A0A2J7PEY7 A0A1I8PCR2 A0A182QRQ8 A0A182NCN7 A0A0A1XI80 A0A0J7N5N7 A0A1Q3F1Z5 B0W3D5 C4WYC0 A0A2A3ETA8 A0A195D1Q6 A0A0L0CQH3 A0A1B6LFV5 A3EXT0 A0A1B6EZH7 A0A0P6H4Q3 A0A1Z5KUS8 A0A1B6I2Q6 A0A0P5HKF2 A0A0P5XC94 A0A0P6B709 E9H9I9 A0A2R5LIA9 A0A0P6JPZ3 A0A0P5ZJB1 A0A293LJ02 A0A131YBB6 A0A0K8RL62 A0A0P5IS99 B7Q8Y0 A0A1B6DD35 L7M7F2 A0A1S3I255 A0A224Z2Q1 A0A0C9R1S0
A0A194PEE9 S4P852 A0A212F720 A0A154P8E9 Q16VF9 A0A195F3D7 A0A1A9VRF1 A0A182S705 A0A182HE04 D3TQW5 A0A151IXT0 A0A2M4BXD9 A0A0L7R232 A0A2P8XXK5 A0A182W325 A0A195BDE7 A0A158NZR6 W5JH54 A0A182IJ22 A0A182YJN2 A0A2M3ZJF1 A0A182MMJ8 A0A151WY04 A0A2M4AXL7 A0A182KEH7 A0A084VFV0 A0A336LL99 A0A1A9WX10 A0A182P3G7 F4WLB6 A0A023EL89 A0A182F772 E2BPM4 A0A088AE86 A0A0L7KN25 A0A1A9XDV8 A0A182R8F6 A0A1B0B8H6 A0A0C9RBL6 A0A1J1HFN6 A0A1I8M9S7 A0A0M9A129 E2A6K8 A0A182UTI9 A0A182LNH4 A0A182HVH7 A0A182XF10 A0A0K8VRH6 A0A232F9F0 A0A026W318 K7IP59 Q7PQ85 A0A034VRH1 A0A067RCZ4 A0A182TT63 A0A1B0G0S1 W8B522 E0VN76 A0A2J7PEY7 A0A1I8PCR2 A0A182QRQ8 A0A182NCN7 A0A0A1XI80 A0A0J7N5N7 A0A1Q3F1Z5 B0W3D5 C4WYC0 A0A2A3ETA8 A0A195D1Q6 A0A0L0CQH3 A0A1B6LFV5 A3EXT0 A0A1B6EZH7 A0A0P6H4Q3 A0A1Z5KUS8 A0A1B6I2Q6 A0A0P5HKF2 A0A0P5XC94 A0A0P6B709 E9H9I9 A0A2R5LIA9 A0A0P6JPZ3 A0A0P5ZJB1 A0A293LJ02 A0A131YBB6 A0A0K8RL62 A0A0P5IS99 B7Q8Y0 A0A1B6DD35 L7M7F2 A0A1S3I255 A0A224Z2Q1 A0A0C9R1S0
PDB
5WLC
E-value=2.74718e-08,
Score=137
Ontologies
GO
PANTHER
Topology
Subcellular location
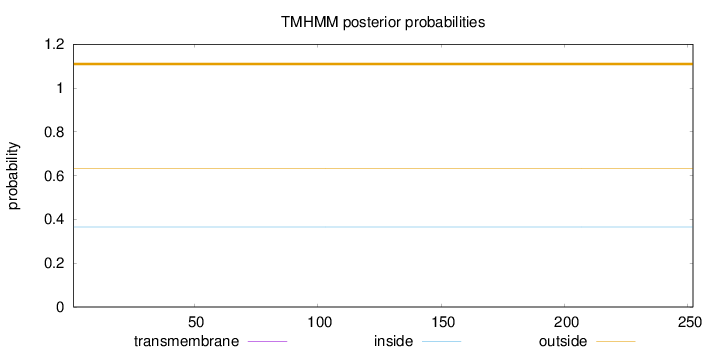
Length:
252
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.36593
outside
1 - 252
Population Genetic Test Statistics
Pi
300.830522
Theta
192.899187
Tajima's D
1.330484
CLR
0.569814
CSRT
0.746362681865907
Interpretation
Uncertain
