Gene
KWMTBOMO07590
Pre Gene Modal
BGIBMGA013701
Annotation
PREDICTED:_E3_ubiquitin-protein_ligase_TM129-like_[Amyelois_transitella]
Full name
E3 ubiquitin-protein ligase TM129
Alternative Name
RING-type E3 ubiquitin transferase TM129
Location in the cell
PlasmaMembrane Reliability : 4.293
Sequence
CDS
ATGGATGTTTTAGTTACACTTTTCTACGTTCTATTCTCTATATGCGTTGTTTATCCGCCTACCGAGTTCGTGTCTGCAGGATTCACAATTGCTCAGCTATTTGATAACTACTTGGGCTCTGAAAATACCAACTTTATTGGTTATCACATGAAGAGAATAACCATAACTGCGTTTATACATTCCGCTCTACCTCTTGGGTACGTTTGCACACTATGGTGTTGCGGGGAACGTAGTCAATGGATGCTTGCAAGTTTTGCTGCGACGGCCTTCATACCGCTGTTAATGTGCTACAAATTGCTGTGCTGGTGGGAGTATGAGAAGCTTGGTCATCCGATCGTAAAGGCACTCCTGCCGTACGTGTCGACGGGAAGTGACTGGCGGGTCACTGCGGCTTATCTCAATTCAGAATTTAGAGGTGTTGATAAAGTCTCAATACAGCTGACGGCTACAAGCAAGTTTGTGGCTACGGAGACTTGGCTCATCAAGGTGTCACAGTATTCCGTTAACATAGTGAAACAGGAAGATTGCTCTCTTGTTGCCACTGCTAGTGACAGCCACAATTTGTCACCATCGGGTGAAGACGTAATCCAATATCTGAACATAGAGGTGATACCCAACCGAGATGAGATCCAGCGATTTACATTTCGCATCACAACAGTAATGTTGCGTGACTTGCAGCCGAGGCTATTGAATGCAGTGAGGGTTCCTGAACACATTTCATTGATGCCAACTTTAGTGGAACGATTCGTGAATGTCTTCAAACAGTATGTTGATCAGAACCCTGTTTATATTATTGATGAGGAGCCAGAGATGTGCATTGGGTGCATGCAAGCCCCGGCTGATGTTAAGATAAATCGACGTTGCTTGCCGCCTCCACCACATCTCGAGGGAGGACCACAGCAGTGCCAGCAGTGTCACTGCAGGTCAGGAAGCAGATATGTCGAATGTGTCGAATTGGCCACGACGAGCTAG
Protein
MDVLVTLFYVLFSICVVYPPTEFVSAGFTIAQLFDNYLGSENTNFIGYHMKRITITAFIHSALPLGYVCTLWCCGERSQWMLASFAATAFIPLLMCYKLLCWWEYEKLGHPIVKALLPYVSTGSDWRVTAAYLNSEFRGVDKVSIQLTATSKFVATETWLIKVSQYSVNIVKQEDCSLVATASDSHNLSPSGEDVIQYLNIEVIPNRDEIQRFTFRITTVMLRDLQPRLLNAVRVPEHISLMPTLVERFVNVFKQYVDQNPVYIIDEEPEMCIGCMQAPADVKINRRCLPPPPHLEGGPQQCQQCHCRSGSRYVECVELATTS
Summary
Description
E3 ubiquitin-protein ligase involved in ER-associated protein degradation, preferentially associates with the E2 enzyme UBE2J2.
Catalytic Activity
S-ubiquitinyl-[E2 ubiquitin-conjugating enzyme]-L-cysteine + [acceptor protein]-L-lysine = [E2 ubiquitin-conjugating enzyme]-L-cysteine + N(6)-ubiquitinyl-[acceptor protein]-L-lysine.
Subunit
Integral component of ER-resident dislocation complexes.
Similarity
Belongs to the TMEM129 family.
Keywords
Complete proteome
Endoplasmic reticulum
Membrane
Metal-binding
Reference proteome
Transferase
Transmembrane
Transmembrane helix
Ubl conjugation pathway
Unfolded protein response
Zinc
Zinc-finger
Feature
chain E3 ubiquitin-protein ligase TM129
Uniprot
H9JVY8
A0A2A4JSA8
A0A2H1X2Y3
A0A212EWX7
A0A194PCW0
A0A194RHR6
+ More
A0A1W4WKD7 A0A067R404 K7IPG7 A0A195BC30 A0A158NN10 A0A232FMR9 A0A195EXW3 A0A151JBF1 A0A151WHH3 A0A195C7J6 F4WT98 E2A9Y5 A0A139WCB1 A0A310SPZ2 A0A1Y1LTC3 A0A3L8DK47 A0A087ZND7 A0A0N0U3M7 A0A2P8Z950 A0A023ERB7 V5I946 A0A182GJQ5 A0A0L7R4G9 A0A2A3E790 A0A1S4F5L7 Q17EJ3 A0A0L8ICQ4 A0A069DTD5 A0A3S1C0F5 A0A2R7WP46 A0A224XEI3 A0A1Q3FHE9 A0A2C9K2B8 A0A026W9D0 F1QX65 A0A0V0G7H2 A0A023F8A8 A0A3M6TSF2 C0HAW5 A0A1S3IYV6 E9H8T9 Q6PD82 A0A060XT69 A0A0P6A465 A0A1B6LAU6 A0A0P4YLJ6 A0A0P5Z7J7 A0A0P5IL30 A0A0P5LS33 A7RGW0 A0A0P6DNM4 A0A0P5QHE3 A0A3P8Y991 G3TMM0 M3WRD8 B0XD82 A0A1A6GXD8 A0A2Y9DZP9 A0A0N7Z905 R4FMN6 A0A0B6ZXK3 A0A336MJS4 Q0IJ33 A0A1U7Q9Q8 A0A3B4CZH3 A0A1A7YPX4 A0A2Y9JM33 L5K4W8 H3B2H5 W5MYU1 I3N2Z2 V4A521 A0A1L8HT77 K9IXG6 A0A2K6JW64 A0A2K6R536 A0A2K6GPX7 E2BEE6 M3YI83 L9KJR7 K1PM91 H0XC45 A0A2K6JW65 A0A2K6R530 Q6IR55 A0A3P4KE28 G9KTT4 A0A1A8S8M6 A0A3Q7SEH5 A0A1A8MB62 F7BAK5
A0A1W4WKD7 A0A067R404 K7IPG7 A0A195BC30 A0A158NN10 A0A232FMR9 A0A195EXW3 A0A151JBF1 A0A151WHH3 A0A195C7J6 F4WT98 E2A9Y5 A0A139WCB1 A0A310SPZ2 A0A1Y1LTC3 A0A3L8DK47 A0A087ZND7 A0A0N0U3M7 A0A2P8Z950 A0A023ERB7 V5I946 A0A182GJQ5 A0A0L7R4G9 A0A2A3E790 A0A1S4F5L7 Q17EJ3 A0A0L8ICQ4 A0A069DTD5 A0A3S1C0F5 A0A2R7WP46 A0A224XEI3 A0A1Q3FHE9 A0A2C9K2B8 A0A026W9D0 F1QX65 A0A0V0G7H2 A0A023F8A8 A0A3M6TSF2 C0HAW5 A0A1S3IYV6 E9H8T9 Q6PD82 A0A060XT69 A0A0P6A465 A0A1B6LAU6 A0A0P4YLJ6 A0A0P5Z7J7 A0A0P5IL30 A0A0P5LS33 A7RGW0 A0A0P6DNM4 A0A0P5QHE3 A0A3P8Y991 G3TMM0 M3WRD8 B0XD82 A0A1A6GXD8 A0A2Y9DZP9 A0A0N7Z905 R4FMN6 A0A0B6ZXK3 A0A336MJS4 Q0IJ33 A0A1U7Q9Q8 A0A3B4CZH3 A0A1A7YPX4 A0A2Y9JM33 L5K4W8 H3B2H5 W5MYU1 I3N2Z2 V4A521 A0A1L8HT77 K9IXG6 A0A2K6JW64 A0A2K6R536 A0A2K6GPX7 E2BEE6 M3YI83 L9KJR7 K1PM91 H0XC45 A0A2K6JW65 A0A2K6R530 Q6IR55 A0A3P4KE28 G9KTT4 A0A1A8S8M6 A0A3Q7SEH5 A0A1A8MB62 F7BAK5
EC Number
2.3.2.27
Pubmed
19121390
22118469
26354079
24845553
20075255
21347285
+ More
28648823 21719571 20798317 18362917 19820115 28004739 30249741 29403074 24945155 26483478 17510324 26334808 15562597 24508170 23594743 25474469 30382153 20433749 21292972 24755649 17615350 25069045 17975172 27129103 23258410 9215903 23254933 27762356 25362486 23385571 22992520 23236062 19892987
28648823 21719571 20798317 18362917 19820115 28004739 30249741 29403074 24945155 26483478 17510324 26334808 15562597 24508170 23594743 25474469 30382153 20433749 21292972 24755649 17615350 25069045 17975172 27129103 23258410 9215903 23254933 27762356 25362486 23385571 22992520 23236062 19892987
EMBL
BABH01036510
NWSH01000725
PCG74578.1
ODYU01013047
SOQ59693.1
AGBW02011879
+ More
OWR45985.1 KQ459606 KPI91062.1 KQ460205 KPJ17107.1 KK852714 KDR17894.1 KQ976529 KYM81750.1 ADTU01020819 NNAY01000011 OXU32031.1 KQ981940 KYN32719.1 KQ979147 KYN22432.1 KQ983115 KYQ47282.1 KQ978231 KYM96136.1 GL888334 EGI62561.1 GL437987 EFN69753.1 KQ971371 KYB25563.1 KQ760287 OAD60880.1 GEZM01048962 JAV76241.1 QOIP01000007 RLU20553.1 KQ435897 KOX69201.1 PYGN01000142 PSN53019.1 GAPW01001878 JAC11720.1 GALX01003570 JAB64896.1 JXUM01068660 KQ562512 KXJ75741.1 KQ414657 KOC65780.1 KZ288357 PBC27152.1 CH477282 EAT44886.1 KQ416074 KOF98810.1 GBGD01001873 JAC87016.1 RQTK01000444 RUS79554.1 KK855191 PTY21364.1 GFTR01005540 JAW10886.1 GFDL01008093 JAV26952.1 KK107347 EZA52281.1 FO904857 FP103003 GECL01002468 JAP03656.1 GBBI01001151 JAC17561.1 RCHS01003041 RMX44259.1 BT059471 ACN11184.1 GL732605 EFX71879.1 BC058876 FR905989 CDQ82497.1 GDIP01035069 JAM68646.1 GEBQ01019150 JAT20827.1 GDIP01225599 JAI97802.1 GDIP01047765 LRGB01000084 JAM55950.1 KZS21072.1 GDIQ01212746 JAK38979.1 GDIQ01190425 JAK61300.1 DS469510 EDO49123.1 GDIQ01075963 JAN18774.1 GDIQ01114094 JAL37632.1 AANG04001605 DS232745 EDS45336.1 LZPO01066269 OBS70826.1 GDKW01001891 JAI54704.1 ACPB03002190 GAHY01001649 JAA75861.1 HACG01026458 CEK73323.1 UFQT01001042 SSX28607.1 BC121210 HADW01015978 HADX01009972 SBP32204.1 KB031030 ELK06590.1 AFYH01076629 AFYH01076630 AFYH01076631 AHAT01001586 AGTP01011815 KB201194 ESO98998.1 CM004466 OCT99238.1 GABZ01006689 JAA46836.1 GL447768 EFN85991.1 AEYP01079003 KB320803 ELW62749.1 JH816697 EKC25087.1 AAQR03169491 BC071045 CYRY02000461 VCW49982.1 JP019715 AES08313.1 HAEH01020137 HAEI01012143 SBS14611.1 HAEF01012893 HAEG01000261 SBR54052.1
OWR45985.1 KQ459606 KPI91062.1 KQ460205 KPJ17107.1 KK852714 KDR17894.1 KQ976529 KYM81750.1 ADTU01020819 NNAY01000011 OXU32031.1 KQ981940 KYN32719.1 KQ979147 KYN22432.1 KQ983115 KYQ47282.1 KQ978231 KYM96136.1 GL888334 EGI62561.1 GL437987 EFN69753.1 KQ971371 KYB25563.1 KQ760287 OAD60880.1 GEZM01048962 JAV76241.1 QOIP01000007 RLU20553.1 KQ435897 KOX69201.1 PYGN01000142 PSN53019.1 GAPW01001878 JAC11720.1 GALX01003570 JAB64896.1 JXUM01068660 KQ562512 KXJ75741.1 KQ414657 KOC65780.1 KZ288357 PBC27152.1 CH477282 EAT44886.1 KQ416074 KOF98810.1 GBGD01001873 JAC87016.1 RQTK01000444 RUS79554.1 KK855191 PTY21364.1 GFTR01005540 JAW10886.1 GFDL01008093 JAV26952.1 KK107347 EZA52281.1 FO904857 FP103003 GECL01002468 JAP03656.1 GBBI01001151 JAC17561.1 RCHS01003041 RMX44259.1 BT059471 ACN11184.1 GL732605 EFX71879.1 BC058876 FR905989 CDQ82497.1 GDIP01035069 JAM68646.1 GEBQ01019150 JAT20827.1 GDIP01225599 JAI97802.1 GDIP01047765 LRGB01000084 JAM55950.1 KZS21072.1 GDIQ01212746 JAK38979.1 GDIQ01190425 JAK61300.1 DS469510 EDO49123.1 GDIQ01075963 JAN18774.1 GDIQ01114094 JAL37632.1 AANG04001605 DS232745 EDS45336.1 LZPO01066269 OBS70826.1 GDKW01001891 JAI54704.1 ACPB03002190 GAHY01001649 JAA75861.1 HACG01026458 CEK73323.1 UFQT01001042 SSX28607.1 BC121210 HADW01015978 HADX01009972 SBP32204.1 KB031030 ELK06590.1 AFYH01076629 AFYH01076630 AFYH01076631 AHAT01001586 AGTP01011815 KB201194 ESO98998.1 CM004466 OCT99238.1 GABZ01006689 JAA46836.1 GL447768 EFN85991.1 AEYP01079003 KB320803 ELW62749.1 JH816697 EKC25087.1 AAQR03169491 BC071045 CYRY02000461 VCW49982.1 JP019715 AES08313.1 HAEH01020137 HAEI01012143 SBS14611.1 HAEF01012893 HAEG01000261 SBR54052.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000192223
+ More
UP000027135 UP000002358 UP000078540 UP000005205 UP000215335 UP000078541 UP000078492 UP000075809 UP000078542 UP000007755 UP000000311 UP000007266 UP000279307 UP000005203 UP000053105 UP000245037 UP000069940 UP000249989 UP000053825 UP000242457 UP000008820 UP000053454 UP000271974 UP000076420 UP000053097 UP000000437 UP000275408 UP000087266 UP000085678 UP000000305 UP000193380 UP000076858 UP000001593 UP000265140 UP000007646 UP000011712 UP000002320 UP000092124 UP000248480 UP000015103 UP000008143 UP000189706 UP000261440 UP000248482 UP000010552 UP000008672 UP000018468 UP000005215 UP000030746 UP000186698 UP000233180 UP000233200 UP000233160 UP000008237 UP000000715 UP000011518 UP000005408 UP000005225 UP000286640 UP000002281
UP000027135 UP000002358 UP000078540 UP000005205 UP000215335 UP000078541 UP000078492 UP000075809 UP000078542 UP000007755 UP000000311 UP000007266 UP000279307 UP000005203 UP000053105 UP000245037 UP000069940 UP000249989 UP000053825 UP000242457 UP000008820 UP000053454 UP000271974 UP000076420 UP000053097 UP000000437 UP000275408 UP000087266 UP000085678 UP000000305 UP000193380 UP000076858 UP000001593 UP000265140 UP000007646 UP000011712 UP000002320 UP000092124 UP000248480 UP000015103 UP000008143 UP000189706 UP000261440 UP000248482 UP000010552 UP000008672 UP000018468 UP000005215 UP000030746 UP000186698 UP000233180 UP000233200 UP000233160 UP000008237 UP000000715 UP000011518 UP000005408 UP000005225 UP000286640 UP000002281
PRIDE
Pfam
PF10272 Tmpp129
Interpro
IPR018801
TM129
ProteinModelPortal
H9JVY8
A0A2A4JSA8
A0A2H1X2Y3
A0A212EWX7
A0A194PCW0
A0A194RHR6
+ More
A0A1W4WKD7 A0A067R404 K7IPG7 A0A195BC30 A0A158NN10 A0A232FMR9 A0A195EXW3 A0A151JBF1 A0A151WHH3 A0A195C7J6 F4WT98 E2A9Y5 A0A139WCB1 A0A310SPZ2 A0A1Y1LTC3 A0A3L8DK47 A0A087ZND7 A0A0N0U3M7 A0A2P8Z950 A0A023ERB7 V5I946 A0A182GJQ5 A0A0L7R4G9 A0A2A3E790 A0A1S4F5L7 Q17EJ3 A0A0L8ICQ4 A0A069DTD5 A0A3S1C0F5 A0A2R7WP46 A0A224XEI3 A0A1Q3FHE9 A0A2C9K2B8 A0A026W9D0 F1QX65 A0A0V0G7H2 A0A023F8A8 A0A3M6TSF2 C0HAW5 A0A1S3IYV6 E9H8T9 Q6PD82 A0A060XT69 A0A0P6A465 A0A1B6LAU6 A0A0P4YLJ6 A0A0P5Z7J7 A0A0P5IL30 A0A0P5LS33 A7RGW0 A0A0P6DNM4 A0A0P5QHE3 A0A3P8Y991 G3TMM0 M3WRD8 B0XD82 A0A1A6GXD8 A0A2Y9DZP9 A0A0N7Z905 R4FMN6 A0A0B6ZXK3 A0A336MJS4 Q0IJ33 A0A1U7Q9Q8 A0A3B4CZH3 A0A1A7YPX4 A0A2Y9JM33 L5K4W8 H3B2H5 W5MYU1 I3N2Z2 V4A521 A0A1L8HT77 K9IXG6 A0A2K6JW64 A0A2K6R536 A0A2K6GPX7 E2BEE6 M3YI83 L9KJR7 K1PM91 H0XC45 A0A2K6JW65 A0A2K6R530 Q6IR55 A0A3P4KE28 G9KTT4 A0A1A8S8M6 A0A3Q7SEH5 A0A1A8MB62 F7BAK5
A0A1W4WKD7 A0A067R404 K7IPG7 A0A195BC30 A0A158NN10 A0A232FMR9 A0A195EXW3 A0A151JBF1 A0A151WHH3 A0A195C7J6 F4WT98 E2A9Y5 A0A139WCB1 A0A310SPZ2 A0A1Y1LTC3 A0A3L8DK47 A0A087ZND7 A0A0N0U3M7 A0A2P8Z950 A0A023ERB7 V5I946 A0A182GJQ5 A0A0L7R4G9 A0A2A3E790 A0A1S4F5L7 Q17EJ3 A0A0L8ICQ4 A0A069DTD5 A0A3S1C0F5 A0A2R7WP46 A0A224XEI3 A0A1Q3FHE9 A0A2C9K2B8 A0A026W9D0 F1QX65 A0A0V0G7H2 A0A023F8A8 A0A3M6TSF2 C0HAW5 A0A1S3IYV6 E9H8T9 Q6PD82 A0A060XT69 A0A0P6A465 A0A1B6LAU6 A0A0P4YLJ6 A0A0P5Z7J7 A0A0P5IL30 A0A0P5LS33 A7RGW0 A0A0P6DNM4 A0A0P5QHE3 A0A3P8Y991 G3TMM0 M3WRD8 B0XD82 A0A1A6GXD8 A0A2Y9DZP9 A0A0N7Z905 R4FMN6 A0A0B6ZXK3 A0A336MJS4 Q0IJ33 A0A1U7Q9Q8 A0A3B4CZH3 A0A1A7YPX4 A0A2Y9JM33 L5K4W8 H3B2H5 W5MYU1 I3N2Z2 V4A521 A0A1L8HT77 K9IXG6 A0A2K6JW64 A0A2K6R536 A0A2K6GPX7 E2BEE6 M3YI83 L9KJR7 K1PM91 H0XC45 A0A2K6JW65 A0A2K6R530 Q6IR55 A0A3P4KE28 G9KTT4 A0A1A8S8M6 A0A3Q7SEH5 A0A1A8MB62 F7BAK5
Ontologies
GO
PANTHER
Topology
Subcellular location
Endoplasmic reticulum membrane
SignalP
Position: 1 - 26,
Likelihood: 0.768802
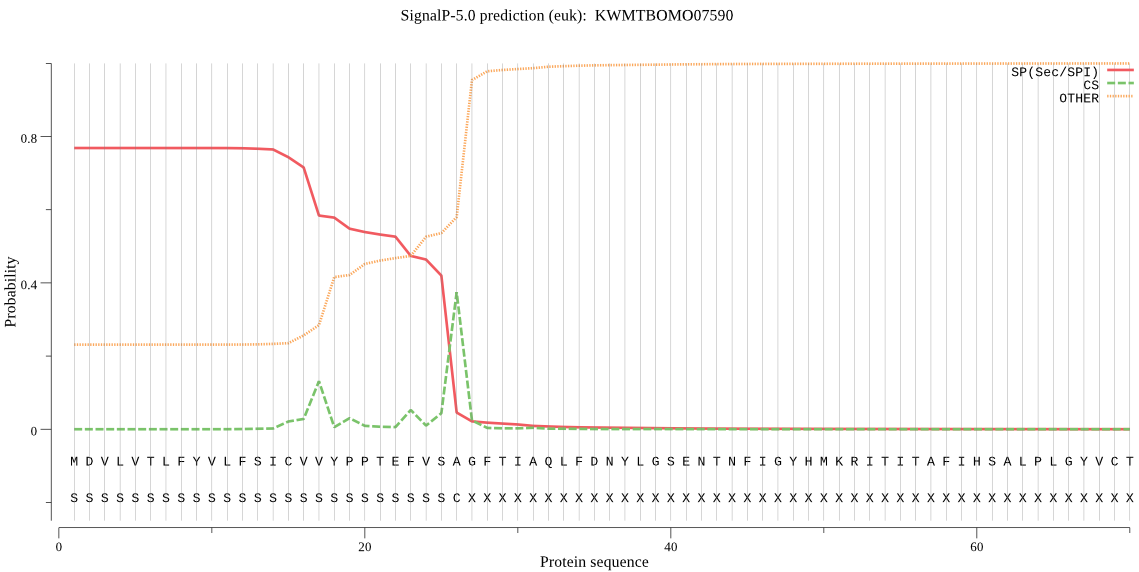
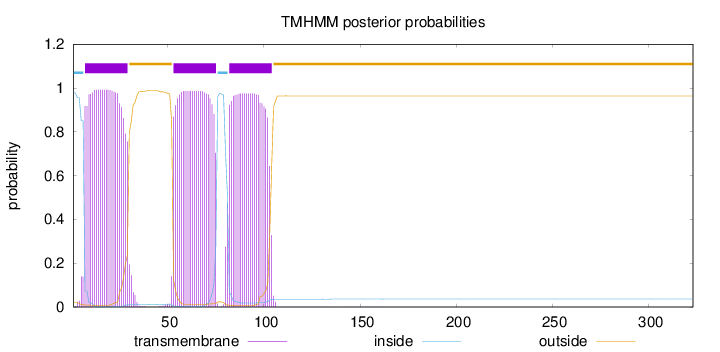
Length:
323
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
66.6856100000002
Exp number, first 60 AAs:
30.54005
Total prob of N-in:
0.98045
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 52
TMhelix
53 - 75
inside
76 - 81
TMhelix
82 - 104
outside
105 - 323
Population Genetic Test Statistics
Pi
195.309573
Theta
172.605867
Tajima's D
0.729759
CLR
0.408219
CSRT
0.576471176441178
Interpretation
Uncertain
