Pre Gene Modal
BGIBMGA013718
Annotation
PREDICTED:_helicase_POLQ-like_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.444
Sequence
CDS
ATGAATTCAAAAGATAAAAGAAACATAGAACATACTTCAACACCTAAACGTCGTCGTCTTTCACTCAGACGAAAATGTTCAAATTTAAAAACATACATTTACAATGCACCTCATGAAGATTCTATACCTGAAATAGCTGATGAAGAAACTAATACGATACACCGATGTGAGTCACCGCGTCTACCGTGTAACCAAGTTGAATATTTGAGTGAATCTGGGAACAGTAGCGATATTCCATGTTCCCCGGGAGTTTCGAATAACTATACCTTAAACAACATCAGTAACTGGGAATCACTAGTATCCAGTGCAAATAACAAGAGTTCTAGTCATATAATTAAGCAAGAAATTGAAAACATATCTGATGAGATGTTCTGTTCGCTGATCGAAAAATCTGTTTCTAATCAAGAAGAAGTTGTTATGAAAAATGATGATAATAATGTTAATAGTCAAGCCAATCACAATCAAATCTTCAAGCTTAACTGTGGTTCCAAAGATGAATTAGAAACTAGAAATGAACTTATGCCAATTTCTCCAATAATAAATAGTAAATCATTTACTTTTAATAAAGGAACTAATCAAAATGAAAATATCAATGCCAGCACTATTAATTATGAAGTTGATATTGAAAATTATTTTGATGAGATCAACCATACCATAAGTAACTTCGATACAGTAAAATTGAATAAATCATCCCTTTTTGAAACAAAAGATTCATTTTTACTTGACATTAAAGATGACAGTATTCTAGGTCAAGAATTCAAATCGGATATTCATGATCTAGTTGAAAGAAAATTAGAAACAAAATTGAATGACAAGCAAGCAGTAATGAGTAATTTGAAAAGTTCAAATAGTTTTTATGGACTCCCGATCTCTGCTAAAGAGCTATTTAAAACTTTTAGGAATATAGAGAGGTTTTATGATTGGCAAGAGGAATGTCTGAATCTAGATGCAATAAAACATAGAAGAAATTTGATATATGCTCTACCAACTAGTGGTGGGAAGACCTTGGTGTCTGAGGTTTTAATGATGAGAGAAGTCATCAACAGGAAGAAGAATGCTTTGTTCATATTACCCTATGTTGCTATTGTTCAGGAAAAAATCTGGGCTCTGTCACCATTTGCAGTGCAGCTAGATTTCCTAATAGAAGAATATGCTGGTGGAAAAGGCAGTCTACCGCCAAAGAAACGACGCAAAAAGAACACTATTTACATTGCGACCATCGAGAAGGGTTTGGCACTTGTTAGAAGTCTCATAGAGCTGCGAAGACTGGACGAACTTGGACTTATTGTGGTAGACGAACTTCATCTTATTGGAGAGAAAGGACGCGGCGGTACTCTAGAGACGTTGTTGACCACTGTCATATTCGTTAACCAGAATATACAAATAGTCGGCATGAGCGCGACGATAGGGAACTTAAGCGAAGTGGCAACGTTCCTGAAGGCGGAAGTGTACGAACGTCACTTCCGGCCGGTGGAGCTGACTGAGTACGTGAAGCTCGGGAGCACGCTGCACAGGATGCTGTGGACAGACGAGGGCTTGGAGCTTGTACCTGATAGAGAGTTGGATTTTAATTACTCGGCGGAGCAGCGCGCGCGCGACCCGGACGGCGTGTGCGGGCTGCTGCGCGGGGGGCGGGCCGCGCTCGTGTTCTGTCCCACGCAGAGGAGCTGCGAGAACGTCGCCGCCACTGTAGCCGCCATGTTGCCGCCAGAGATGGCAGACGGTCGCGCCGAGGAACGCTTAGCTCTGGTTGAAGCTCTGAGGGCCGAGGGCTCCACCGACGCCCTGCTGCAGGCGGCGTCCTGCGGCGTGGCCTACCACCACGCGGGGCTTGCGACCGACGAGCGACACTTGTTGGAACAGGCCTTCAGGTCGGGGGTGCTGTCCACCATATGCTGCACGTCGACCCTGGCGGCCGGCGTGAACCTGCCGGCGGCGCGCGTGCTGCTGCGGGGCGCGGCGGGCGCGCGGGGGGGGCCGCCCGCGCTCTCCGCCGCCGCCTACCGACAGATGGCGGGCCGGGCGGGCAGGGCGGGGCTCGCCGCCGCCGGCGAAAGCATACTAATATGCAGCCCCCAGCAGTGGGAGCATTTGAAGCCAGTACTGGAAGCGGGAGTGGCCCCGGTGACCAGCGTCCTGCGACCCTCGCTAGAGAGCTTAATACTGAGCGCCGTGACTCTACAGTTAGCGGACACTGGCCCCGAGCTGAAGAGACTGCTGGACTCCACGTTCATGGCTGTCCCGAAAGCTGCTGAAAGCGCGGCCGCCGACACGAGGGCGGCGTGCCAGGAGGTGCTGCGGTCCCTCACGCGTGCCGGAGTCCTGGACGTTAAGGGCCAAAGGAAACTAAACCAGGACAATGATGATGAGGAGCTATACAACTCTCAGTTCGTCGTCAGCAGACTCGGTCAAGCCGCTGTCAAAGGATGCCTGTCGGTGGAGGCGGCGCGGCGGCTGGTGGCGGAGCTGCGTCGTGCGGCGCGCGGGCTGGTGCTGTGCGGGCCGCTGCACCTGCTGTACCTGGCGGCGCCGCACGACGACCCCGTGCGGCCCGACCCGAGACACTTTTACTCGCTGTACTGCGAACTAGACGAAGAGGGCCTGCAGACAGCTAAGGCGCTGGGCATCAACGAGACGAACGCGATCCGCATGATGACCGGGAGACCCATGGCTTCCGTGCCGTCGCTGGTCGTGAGCCGGTTCTACATGGCGCTGATGCTGCGCGATCTCTGGCGACACATGCCGCTGCACGCCGTGGCCGACAATGCTCAGGCCGTGTACTCGGTGCAGATACCTGGTGGGGCGGGGCCGCGTGCAGTGGCTGCTGACGGCGGGGTGCGCGGGGGCGGGGGGCGCGGCGGCGGCGTGCGAGGCGCTGGGGGCGGCGCCCTTCGCCGCGCTGCTGCAGCCGCTGGCGCTGGCGCTCAGGACGTGCGCTCATCCGCAACTACACCTGCTCATGGAACTGCCAGCCGTGAAAAAAGCCCGTGCGCTGCAGCTGATGCGTGCGGGATACAAGAGAATAGAAGACGTCGCTAA
Protein
MNSKDKRNIEHTSTPKRRRLSLRRKCSNLKTYIYNAPHEDSIPEIADEETNTIHRCESPRLPCNQVEYLSESGNSSDIPCSPGVSNNYTLNNISNWESLVSSANNKSSSHIIKQEIENISDEMFCSLIEKSVSNQEEVVMKNDDNNVNSQANHNQIFKLNCGSKDELETRNELMPISPIINSKSFTFNKGTNQNENINASTINYEVDIENYFDEINHTISNFDTVKLNKSSLFETKDSFLLDIKDDSILGQEFKSDIHDLVERKLETKLNDKQAVMSNLKSSNSFYGLPISAKELFKTFRNIERFYDWQEECLNLDAIKHRRNLIYALPTSGGKTLVSEVLMMREVINRKKNALFILPYVAIVQEKIWALSPFAVQLDFLIEEYAGGKGSLPPKKRRKKNTIYIATIEKGLALVRSLIELRRLDELGLIVVDELHLIGEKGRGGTLETLLTTVIFVNQNIQIVGMSATIGNLSEVATFLKAEVYERHFRPVELTEYVKLGSTLHRMLWTDEGLELVPDRELDFNYSAEQRARDPDGVCGLLRGGRAALVFCPTQRSCENVAATVAAMLPPEMADGRAEERLALVEALRAEGSTDALLQAASCGVAYHHAGLATDERHLLEQAFRSGVLSTICCTSTLAAGVNLPAARVLLRGAAGARGGPPALSAAAYRQMAGRAGRAGLAAAGESILICSPQQWEHLKPVLEAGVAPVTSVLRPSLESLILSAVTLQLADTGPELKRLLDSTFMAVPKAAESAAADTRAACQEVLRSLTRAGVLDVKGQRKLNQDNDDEELYNSQFVVSRLGQAAVKGCLSVEAARRLVAELRRAARGLVLCGPLHLLYLAAPHDDPVRPDPRHFYSLYCELDEEGLQTAKALGINETNAIRMMTGRPMASVPSLVVSRFYMALMLRDLWRHMPLHAVADNAQAVYSVQIPGGAGPRAVAADGGVRGGGGRGGGVRGAGGGALRRAAAAAGAGAQDVRSSATTPAHGTASREKSPCAAADACGIQENRRRR
Summary
Uniprot
EMBL
NWSH01000725
PCG74592.1
RSAL01000208
RVE44274.1
KQ460205
KPJ17101.1
+ More
KQ459606 KPI91056.1 GEZM01057476 JAV72317.1 APGK01039350 KB740970 ENN76644.1 KB632388 ERL94373.1 CH480815 EDW40654.1 CM002912 KMY98290.1 LBMM01000539 KMQ98107.1 JXJN01001784 CCAG010017308 KZ288293 PBC29055.1 GL761987 EFZ22552.1 KQ981727 KYN36744.1 KQ977791 KYM99877.1 CP012525 ALC43373.1
KQ459606 KPI91056.1 GEZM01057476 JAV72317.1 APGK01039350 KB740970 ENN76644.1 KB632388 ERL94373.1 CH480815 EDW40654.1 CM002912 KMY98290.1 LBMM01000539 KMQ98107.1 JXJN01001784 CCAG010017308 KZ288293 PBC29055.1 GL761987 EFZ22552.1 KQ981727 KYN36744.1 KQ977791 KYM99877.1 CP012525 ALC43373.1
Proteomes
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
PDB
5AGA
E-value=7.42781e-55,
Score=545
Ontologies
GO
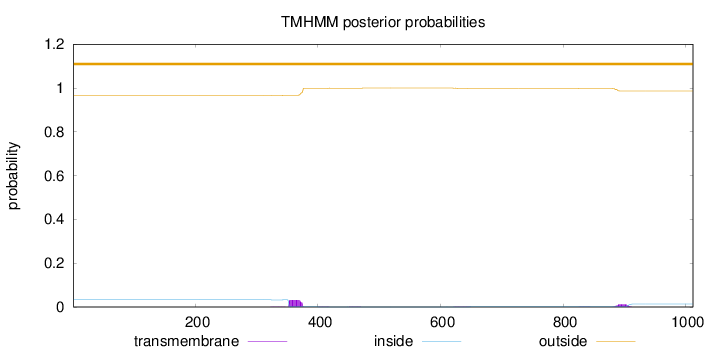
Topology
Length:
1012
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.00378
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03334
outside
1 - 1012
Population Genetic Test Statistics
Pi
209.65911
Theta
149.786057
Tajima's D
1.011168
CLR
1.077932
CSRT
0.664916754162292
Interpretation
Uncertain
