Pre Gene Modal
BGIBMGA013699
Annotation
PREDICTED:_ribosomal_RNA-processing_protein_8_[Plutella_xylostella]
Full name
Ribosomal RNA-processing protein 8
Location in the cell
Mitochondrial Reliability : 2.638
Sequence
CDS
ATGTTCAAAATACCGGAATGGGATAGTTATGTGCCCAAATCAAATATTAAATTCAGTAGGAAAACCGTCAAGGCAAAATCACAAAACTTAATACTGAAAACTGAAACTCCAATAAATATTGAATTAACCAACACCAAGCAGTTTCCAAAAAGTGCCAATAAAAATGATGATCAAGAAGATATTTTATTTAAAGAAAGCAAAGGAAAGAAAGCAAAACACCTTAAAACCAGCAAGGATGAAAATATAAATAAATTAGAAGCATCAAATTATGTTAAAATTGATTTTAAAAAAGAAAAACTAAGAAAGATGTTACAAGAAAACTCATTGAGAAGTCGTTTTACTGTTAATGTGAGAAACGGAAACAAGTTGAGGGAGAGAATGCTTGAAAGATTAAAAGCTGCACAATTCAGGTACCTCAATGAAAAACTATACACATCATCAGGTACAGAAGCTCATCAATTATTCCAGAGTGATCCCGCTGCCTTCAAAACTTACCATGAAGGTTACCAGCAACAACTCAAGAAGTGGCCCATCAATCCATTAGACCTAATTGTAAAGAGGATTCAGAAAATGCCTAAAACACATAAAATAGCTGATATGGGTTGTGGAGAAGCTGCGCTGTCACATCGTGTACCCCATCCAGTTCGGTCCTTTGATTTGGTAGCCACTACACCAAGTGTAGAAGTGTGTAACATAGCACATACCCCACTGCTGGCAGCGTCCATGGATGTTGCAGTCTATTGTTTAGCACTAATGGGCACTGAACTCACTCAGTACTTGGTCGAAGCTAGTAGAATATTGAAAATTGGAGGCCATCTTCTAATAGCAGAAGTTGAGAGTCGTTTTGAAAAAGTTGAGGACTTTACTAAAGCAGTAGAGCGGATCGGATTCAAACTAATTAAACTGGACAAGAGTCACACCGTCTTCTACTTCATGGAGTTCACGAAAATACGAGACGCGCCCGTGAAGAAATCCAAACTACCCGCTTTGACGCTTAAACCTTGTCTTTATAAGAGACGATAA
Protein
MFKIPEWDSYVPKSNIKFSRKTVKAKSQNLILKTETPINIELTNTKQFPKSANKNDDQEDILFKESKGKKAKHLKTSKDENINKLEASNYVKIDFKKEKLRKMLQENSLRSRFTVNVRNGNKLRERMLERLKAAQFRYLNEKLYTSSGTEAHQLFQSDPAAFKTYHEGYQQQLKKWPINPLDLIVKRIQKMPKTHKIADMGCGEAALSHRVPHPVRSFDLVATTPSVEVCNIAHTPLLAASMDVAVYCLALMGTELTQYLVEASRILKIGGHLLIAEVESRFEKVEDFTKAVERIGFKLIKLDKSHTVFYFMEFTKIRDAPVKKSKLPALTLKPCLYKRR
Summary
Description
Probable methyltransferase required to silence rDNA.
Similarity
Belongs to the methyltransferase superfamily. RRP8 family.
Uniprot
EC Number
2.1.1.-
Pubmed
EMBL
BABH01036512
NWSH01000725
PCG74593.1
ODYU01013048
SOQ59695.1
KQ460205
+ More
KPJ17100.1 KQ459606 KPI91054.1 KZ288293 PBC29056.1 BT126684 AEE61646.1 KQ435898 KOX69176.1 QOIP01000001 RLU26956.1 KK107447 EZA50748.1 KB632388 ERL94374.1 APGK01039350 KB740970 ENN76645.1 ADTU01013343 KQ976401 KYM92446.1 KQ978625 KYN29641.1 GDHC01002415 JAQ16214.1 KQ982080 KYQ60370.1 GGFJ01006538 MBW55679.1 CH477399 EAT41758.1 GANP01004013 JAB80455.1
KPJ17100.1 KQ459606 KPI91054.1 KZ288293 PBC29056.1 BT126684 AEE61646.1 KQ435898 KOX69176.1 QOIP01000001 RLU26956.1 KK107447 EZA50748.1 KB632388 ERL94374.1 APGK01039350 KB740970 ENN76645.1 ADTU01013343 KQ976401 KYM92446.1 KQ978625 KYN29641.1 GDHC01002415 JAQ16214.1 KQ982080 KYQ60370.1 GGFJ01006538 MBW55679.1 CH477399 EAT41758.1 GANP01004013 JAB80455.1
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
PDB
2ZFU
E-value=5.17163e-56,
Score=550
Ontologies
GO
PANTHER
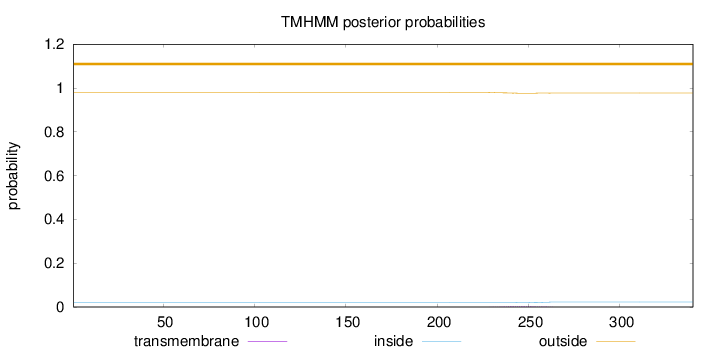
Topology
Subcellular location
Length:
340
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.07
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02062
outside
1 - 340
Population Genetic Test Statistics
Pi
207.880285
Theta
177.269045
Tajima's D
0.367255
CLR
0
CSRT
0.475876206189691
Interpretation
Uncertain
