Gene
KWMTBOMO07585
Pre Gene Modal
BGIBMGA013719
Annotation
PREDICTED:_uncharacterized_protein_LOC101741912_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.872
Sequence
CDS
ATGGGTTACTCTGTGGTCAAAAGATCCATCATCAAAGGGCGGAGGGACAAAATTTGGAACTGGCGCTGCCGATGGAGCATTGTGACGATAGCGGCCATAGCGCTAATATTCTACAACGCCGTTGCCAACATCTGGTTGTTGCATCCCACCGCTTGTCCCGTCATCTCAACTGCCTCTCCCTCAGATCCTCCATCGTGCGAACCATGCATAGACACGGTACCGAATAGCGCAATCGACGACGATCCGATCGCGAAACTCGACTTGCGATTAGGAAGGTGGGACGGATCTCGAAGCTACAGACTCTTCGATTACGCGGCTGTGGGAGAGCTATACTCTGAAGTATCAAGCAACAGTAAGGTGTGCCTCGCCACTCAGAGCTCCATCGAACGCCTCCACGAGTTGCTAAGAATAGCCGCGCACTGGACCGGACCTATATCAGTGGCTGTGTTTGTCGCTGGAGATGAACTAAGACTTTTGAGAGCGTTCACGACATGGCTATTTCGATGTCAACCAGAAATATATTCAAGAGTTGCCTTACATATTGCGATGCCAGCAGAAAGACCAGGAGTGCACGGTGTAGTACCGAGCTGGGCAAGGGATTGCCGCTCGAAGCCGTTGCCTCCAGGAGAAAGAAGGGCAGATACAGTAGCTTGGAGGGCACGACATCCATACCCTCAGAACCATATGAGGAATCAAGCACGGAAGAACTGTCATACTTCTTACGTTTTTCTTGTCGATGTGGATATCGTGCCGTCTAAGGGGATGGCAGCATCGCTCGATAAGTTCTTGACAACAGCACCTAAATGCTCGCTCTGTGCCTATGTTGTTCCGACATATGAACTAGACAAACGCGTGGCGACCTTCCCTAGCAGCAAGTCAGAACTTTTACGCCTCTCCAGAAATAAATTAGCCATTCCGTTTCATAGAAAAGTGTTTATTTACAACCAGTACGCTTCTAATTTTTCTAGATGGGAGGCGAGTGGAGGCAACGAGACCAGCGAGACCCACATCTCCCATAACGTGACCAACTTCGAGCTGTTGTACGAGCCCTTCTACGTGGCTCCGGACACTGTACCTGCTCATGATGAACGGTTTCTGGGCTACGGCTTCACGAGGAATACACAGGAGTGCCGGAAATATACACCAGACCTCCAAGAAGCAGTTGAAGGCGACTGCTGCTCTGATGCTCAGAACACCGATGATGTACAGGAAGATCAGGGCCTGTTAGAGTTGGAAGCGGCTTGTCCAGAGTTGAGCGGACAGATCATAGGTGGGAGACCGAGCTCTGTCAGCAGACACCCTTATCAGGTGTCGATGGTGCTGAACGGTAACTCTTTCTGTGGAGGCTTCATCATCAGCAGAGACTTCGTGCTAACGGCCGCCCATTGTGTTCAGAATACGGTTCCAAGCGCTATTCGCCTTAGGGTTGGCAGCACACGACGTGACTCCGGTGGCAGGATAGTCAGGGTGTCCAATGTGACTGTCCATCCACAGTACGGAAACCCGCAGTTTGACAATGACATAGCTGTGCTCCGCTTGGCCCAGCCGTTGCCGGTCAATGCTGCGATACGGCCTATTAGATTGCCACAGAGAGGCCAGGCTGTGCCTCTCGTCCGATTGACTGTCACTGGATGGGGATTGACTGCTCCTCGTGGTCGAAGAATTCCTCGTATAATGATGGAGGCTCAAGTACCAGTTGTGCCTCACTGGTTGTGTCGCTTGTCCTATGGTGATGCGTTGACCGGGAGTATGTTTTGTGGCGGCCATTTTCTTATTGGAGGAGTATCATCCTGTCAGGGTGATTCCGGAGGTCCAGCAGTCTTCAGAGGGACAGCGTTCGGCATTGTATCATTCGCCAGAGGTTGCGCCCTTCCATTGTCCCCCACCGTGTTCACCAACACAGCGGCCCTTAGAGATTGGATCACGGATAACACTGGAGTTTGA
Protein
MGYSVVKRSIIKGRRDKIWNWRCRWSIVTIAAIALIFYNAVANIWLLHPTACPVISTASPSDPPSCEPCIDTVPNSAIDDDPIAKLDLRLGRWDGSRSYRLFDYAAVGELYSEVSSNSKVCLATQSSIERLHELLRIAAHWTGPISVAVFVAGDELRLLRAFTTWLFRCQPEIYSRVALHIAMPAERPGVHGVVPSWARDCRSKPLPPGERRADTVAWRARHPYPQNHMRNQARKNCHTSYVFLVDVDIVPSKGMAASLDKFLTTAPKCSLCAYVVPTYELDKRVATFPSSKSELLRLSRNKLAIPFHRKVFIYNQYASNFSRWEASGGNETSETHISHNVTNFELLYEPFYVAPDTVPAHDERFLGYGFTRNTQECRKYTPDLQEAVEGDCCSDAQNTDDVQEDQGLLELEAACPELSGQIIGGRPSSVSRHPYQVSMVLNGNSFCGGFIISRDFVLTAAHCVQNTVPSAIRLRVGSTRRDSGGRIVRVSNVTVHPQYGNPQFDNDIAVLRLAQPLPVNAAIRPIRLPQRGQAVPLVRLTVTGWGLTAPRGRRIPRIMMEAQVPVVPHWLCRLSYGDALTGSMFCGGHFLIGGVSSCQGDSGGPAVFRGTAFGIVSFARGCALPLSPTVFTNTAALRDWITDNTGV
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
Pubmed
EMBL
Proteomes
Pfam
PF00089 Trypsin
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
PDB
2VU8
E-value=3.91201e-35,
Score=373
Ontologies
PATHWAY
GO
Topology
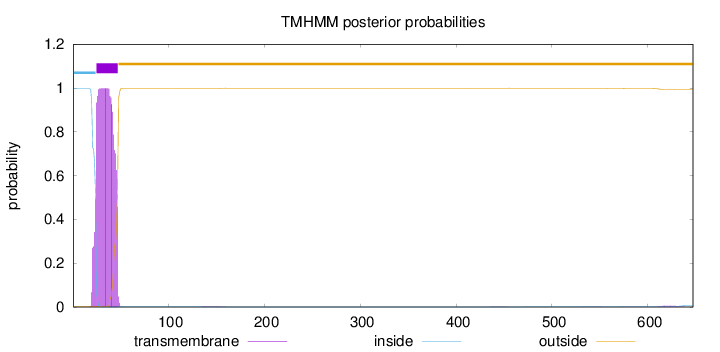
Length:
647
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.6242699999999
Exp number, first 60 AAs:
22.41685
Total prob of N-in:
0.99690
POSSIBLE N-term signal
sequence
inside
1 - 24
TMhelix
25 - 47
outside
48 - 647
Population Genetic Test Statistics
Pi
30.029999
Theta
226.812649
Tajima's D
-1.388912
CLR
0.75467
CSRT
0.0769461526923654
Interpretation
Uncertain
