Gene
KWMTBOMO07578
Pre Gene Modal
BGIBMGA013723
Annotation
PREDICTED:_transmembrane_and_TPR_repeat-containing_protein_1-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.813
Sequence
CDS
ATGGACATCGCAGTATCAATGCCGCTTCTGATCTTGATACATGAGGTGGCCGGTGTGGTGGGGCGTGCAGACGTGTTAGCCTGTGTCTTCTTTCTCACATCTATTCTTCTTTATCACGGAACAAACAATCACCGTGTCTGGGGGAGTGTAGCAGCTGGTGCCCTTAGCATGTTGGCCAAGGAGACAGGACTCACTGCCTTACTCTTCAATGCTGCACTGGACCTGTACAGATGTTGGGCTCATGTGCACAGATCTTCGTCAGCAGTAAAATGGCGTTGGAAAGACAGTCCTTACGGCAGAGCATTTAAAACTCTAGCAGCTCTCAGTTTACTGGCTGGAGCAAGACTAGCATTACTACAAGGTTCCTTACCGGCTTTTTCACCACAGGACAATCCACCTGCTTTCCATCCTTCATTTGCTGTCAGGTTGATGACCTTTTGCTACTTGGCCGCTTTCAACTGGTGGCTGCTAGTGTGTCCATGGACTTTAAGTCACGATTGGCAGATGGGCTCAGTACCATTGATCACCAACGTCAGCGATCTAAGGAATTTAGTCACCTGTGCTGCCATGTGCGCTTTGTTTGGTCTCTGTTACAGATGTCTGCTCGATTTGGAGATGCAGCGTCATACTCCAGCGGTTGTGGGATTAATGCTGCTCGTCATTCCATATCTCCCTGCCTCCAACCTGCTTGTCACTGTGGGTTTCGTCATCGCTGAACGAGTGTTGTATATTCCCAGTGTGGGCTCAGTATTGTTGACTGCGTATGGAATTGAACTGATGTGGAAGCGAGGGTTTAGAACGAAGAAGCTGCTAGTGCTATGCGTTGCAACACTAGTGGCTGCAGGAGTAGCGAAAACACATCTTAGAAACAGAGATTGGAGAACTAGAGAATCTTTACTAAGAGCTGATCTCTCAGTGTTGCCACATAACGCAAAATTACACTACAATTTCGCTAATTTCTTGAAGGACGTCGAGCAGCACGAGATTGCCATCAAACATTACAAGGAAGCATTGAAATTGTGGCCAAGTTACGCGAGTGCCCACAATAACTTGGGCACGCTGGTGCTGGCATCGGGGAGGGCGGAGCACCACTTCCTTCAAGCCCTTAAATACAACAAAGACCACGTTAACGCTCATTACAATCTCGCTAAATTATACAGAAAGAAGAATAGAATACCGGAGGCTCTTAGGATGTTGGAACGTTGCATAACTTTAGAACCAAGATTCGTTCAAGCATATCTGGAACTGTTACTGATGAAACCTGATGAAGAGAAAAGGTGGATCTTGAACAAACTGCTAGAGTTAGAGCCCTACAACTGGGAGCACTATCTTCTTTATGGAAACTGGCTGAGAGGACGAGGTCTTCTCCCGTTATCACTAGAATACTATAGAACAGCGATGCGTCTGTCATTCAGTACGCGACACAACAGTCCGAGGGGCGAGTTACTGTCGCTGCGAGCTGCATGCCTGGCGCTACGGGACACCGGCCAGTACGCGAGGATGCTGCAGTTATTAGTCAGATGGCACGCAGGTCGGGGCGGGCAGCCAGCCGGGGCTCGGCTGCTGGCGGCGCGGGCCTGGATTCTTCGCTCCCAGCTGGCGCGGCGTGCGGCTCTCTACTCCCACACGCATCGACAGATGACATCAACATGCCTACACCACAGTCAACTGGAGGTGTCGCCGGAATTATACATAGATTTTAAACAACAAGACGTCAAACCTAGTTCAAGGCTACACGCGCACGACGCGAAACTTCCCACCACTAATAGCGTAGAGAAGAATTGCCAAAAAACCCACGAAAAAAGTGTGCTCTCAGCCCAGTACAAACCGACTCATAATAAATCTGAAACAGGTCATAAATTGAAAAGTTGTCCTCTGAATAAAAAGAAGGAACGTCGAATAAAAGATCCGGAGACGTTTCCATCTTTTGTTTCGGATCATTTAATTAAGACGTATTAA
Protein
MDIAVSMPLLILIHEVAGVVGRADVLACVFFLTSILLYHGTNNHRVWGSVAAGALSMLAKETGLTALLFNAALDLYRCWAHVHRSSSAVKWRWKDSPYGRAFKTLAALSLLAGARLALLQGSLPAFSPQDNPPAFHPSFAVRLMTFCYLAAFNWWLLVCPWTLSHDWQMGSVPLITNVSDLRNLVTCAAMCALFGLCYRCLLDLEMQRHTPAVVGLMLLVIPYLPASNLLVTVGFVIAERVLYIPSVGSVLLTAYGIELMWKRGFRTKKLLVLCVATLVAAGVAKTHLRNRDWRTRESLLRADLSVLPHNAKLHYNFANFLKDVEQHEIAIKHYKEALKLWPSYASAHNNLGTLVLASGRAEHHFLQALKYNKDHVNAHYNLAKLYRKKNRIPEALRMLERCITLEPRFVQAYLELLLMKPDEEKRWILNKLLELEPYNWEHYLLYGNWLRGRGLLPLSLEYYRTAMRLSFSTRHNSPRGELLSLRAACLALRDTGQYARMLQLLVRWHAGRGGQPAGARLLAARAWILRSQLARRAALYSHTHRQMTSTCLHHSQLEVSPELYIDFKQQDVKPSSRLHAHDAKLPTTNSVEKNCQKTHEKSVLSAQYKPTHNKSETGHKLKSCPLNKKKERRIKDPETFPSFVSDHLIKTY
Summary
Uniprot
A0A2H1VQ98
A0A2A4JI66
A0A2A4JJL1
A0A212EMG9
A0A0N1IEF5
A0A194PIY1
+ More
A0A194RHT7 A0A194PDJ1 A0A3S2LRN5 A0A2H1V1T9 A0A0L7LR60 A0A336KPG1 D6WAH1 A0A087ZQP9 A0A3L8E3Q0 A0A2C9H6G0 A0A1B6IMY3 E0VKZ8 A0A1J1J8M9 J9KBD8 A0A2S2NY28 A0A2H8TL04 A0A2S2QZX7 A0A3B3CL08 A0A1A8CWI6 Q4SZ08 A0A1A8U0B2 A0A1A8MM39 A0A1A8QWS5 A0A1A8FI83 A0A1S3SGI6 A0A146VID1 A0A1A8IGM2 A0A146Q8K5 A0A146PTH2 A0A146VHJ9 A0A1S3MZK1 A0A146ZS10 A0A3P9CVB1 A0A3Q3KLT7 A0A3Q3N444 A0A1A7YQD9 A0A3B1KBM3 A0A1A8M4L0 A0A2D0T657 A0A3P8PZN6 A0A2Y9REW1 F1QF84 T1J4S4 A0A1S3H6C7 A0A3P9IBC1 H2MCF2 A0A3P9PA00 A0A3P8UWG6 A0A3P8UWH1 V4CNL0 A0A147BMK6 A0A3P9LE65 A0A3B3REW5 A0A341AGN4 A0A2J8WC27 A0A195ED44 A0A2I4C0J8 A0A3Q3BFX4
A0A194RHT7 A0A194PDJ1 A0A3S2LRN5 A0A2H1V1T9 A0A0L7LR60 A0A336KPG1 D6WAH1 A0A087ZQP9 A0A3L8E3Q0 A0A2C9H6G0 A0A1B6IMY3 E0VKZ8 A0A1J1J8M9 J9KBD8 A0A2S2NY28 A0A2H8TL04 A0A2S2QZX7 A0A3B3CL08 A0A1A8CWI6 Q4SZ08 A0A1A8U0B2 A0A1A8MM39 A0A1A8QWS5 A0A1A8FI83 A0A1S3SGI6 A0A146VID1 A0A1A8IGM2 A0A146Q8K5 A0A146PTH2 A0A146VHJ9 A0A1S3MZK1 A0A146ZS10 A0A3P9CVB1 A0A3Q3KLT7 A0A3Q3N444 A0A1A7YQD9 A0A3B1KBM3 A0A1A8M4L0 A0A2D0T657 A0A3P8PZN6 A0A2Y9REW1 F1QF84 T1J4S4 A0A1S3H6C7 A0A3P9IBC1 H2MCF2 A0A3P9PA00 A0A3P8UWG6 A0A3P8UWH1 V4CNL0 A0A147BMK6 A0A3P9LE65 A0A3B3REW5 A0A341AGN4 A0A2J8WC27 A0A195ED44 A0A2I4C0J8 A0A3Q3BFX4
Pubmed
EMBL
ODYU01003801
SOQ43021.1
NWSH01001302
PCG71785.1
PCG71784.1
AGBW02013845
+ More
OWR42669.1 KQ460545 KPJ14077.1 KQ459606 KPI91045.1 KQ460205 KPJ17127.1 KPI91078.1 RSAL01000288 RVE42938.1 ODYU01000286 SOQ34779.1 JTDY01000277 KOB77947.1 UFQS01000686 UFQT01000686 SSX06207.1 SSX26561.1 KQ971312 EEZ97998.1 QOIP01000001 RLU27370.1 GECU01019439 JAS88267.1 DS235255 EEB14054.1 CVRI01000074 CRL08148.1 ABLF02040630 ABLF02040638 ABLF02040658 ABLF02040661 GGMR01009500 MBY22119.1 GFXV01002924 MBW14729.1 GGMS01014108 MBY83311.1 HADZ01019095 SBP83036.1 CAAE01011868 CAF94124.1 HADY01024443 HAEJ01000559 SBS41016.1 HAEF01016320 SBR57479.1 HAEH01013764 SBR98250.1 HAEB01012011 HAEC01002462 SBQ58538.1 GCES01069596 JAR16727.1 HAED01010069 SBQ96281.1 GCES01134206 JAQ52116.1 GCES01139171 JAQ47151.1 GCES01069595 JAR16728.1 GCES01017616 JAR68707.1 HADW01005313 HADX01010166 SBP32398.1 HAEF01011145 SBR51747.1 CT573126 CU861462 FO681318 JH431849 KB199905 ESP03985.1 GEGO01003377 JAR92027.1 NDHI03003395 PNJ67291.1 KQ979074 KYN23031.1
OWR42669.1 KQ460545 KPJ14077.1 KQ459606 KPI91045.1 KQ460205 KPJ17127.1 KPI91078.1 RSAL01000288 RVE42938.1 ODYU01000286 SOQ34779.1 JTDY01000277 KOB77947.1 UFQS01000686 UFQT01000686 SSX06207.1 SSX26561.1 KQ971312 EEZ97998.1 QOIP01000001 RLU27370.1 GECU01019439 JAS88267.1 DS235255 EEB14054.1 CVRI01000074 CRL08148.1 ABLF02040630 ABLF02040638 ABLF02040658 ABLF02040661 GGMR01009500 MBY22119.1 GFXV01002924 MBW14729.1 GGMS01014108 MBY83311.1 HADZ01019095 SBP83036.1 CAAE01011868 CAF94124.1 HADY01024443 HAEJ01000559 SBS41016.1 HAEF01016320 SBR57479.1 HAEH01013764 SBR98250.1 HAEB01012011 HAEC01002462 SBQ58538.1 GCES01069596 JAR16727.1 HAED01010069 SBQ96281.1 GCES01134206 JAQ52116.1 GCES01139171 JAQ47151.1 GCES01069595 JAR16728.1 GCES01017616 JAR68707.1 HADW01005313 HADX01010166 SBP32398.1 HAEF01011145 SBR51747.1 CT573126 CU861462 FO681318 JH431849 KB199905 ESP03985.1 GEGO01003377 JAR92027.1 NDHI03003395 PNJ67291.1 KQ979074 KYN23031.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000053268
UP000283053
UP000037510
+ More
UP000007266 UP000005203 UP000279307 UP000075920 UP000009046 UP000183832 UP000007819 UP000261560 UP000087266 UP000265000 UP000265160 UP000261600 UP000261640 UP000018467 UP000221080 UP000265100 UP000248480 UP000000437 UP000085678 UP000265200 UP000001038 UP000242638 UP000265120 UP000030746 UP000265180 UP000261540 UP000252040 UP000078492 UP000192220 UP000264800
UP000007266 UP000005203 UP000279307 UP000075920 UP000009046 UP000183832 UP000007819 UP000261560 UP000087266 UP000265000 UP000265160 UP000261600 UP000261640 UP000018467 UP000221080 UP000265100 UP000248480 UP000000437 UP000085678 UP000265200 UP000001038 UP000242638 UP000265120 UP000030746 UP000265180 UP000261540 UP000252040 UP000078492 UP000192220 UP000264800
Interpro
SUPFAM
SSF48452
SSF48452
Gene 3D
ProteinModelPortal
A0A2H1VQ98
A0A2A4JI66
A0A2A4JJL1
A0A212EMG9
A0A0N1IEF5
A0A194PIY1
+ More
A0A194RHT7 A0A194PDJ1 A0A3S2LRN5 A0A2H1V1T9 A0A0L7LR60 A0A336KPG1 D6WAH1 A0A087ZQP9 A0A3L8E3Q0 A0A2C9H6G0 A0A1B6IMY3 E0VKZ8 A0A1J1J8M9 J9KBD8 A0A2S2NY28 A0A2H8TL04 A0A2S2QZX7 A0A3B3CL08 A0A1A8CWI6 Q4SZ08 A0A1A8U0B2 A0A1A8MM39 A0A1A8QWS5 A0A1A8FI83 A0A1S3SGI6 A0A146VID1 A0A1A8IGM2 A0A146Q8K5 A0A146PTH2 A0A146VHJ9 A0A1S3MZK1 A0A146ZS10 A0A3P9CVB1 A0A3Q3KLT7 A0A3Q3N444 A0A1A7YQD9 A0A3B1KBM3 A0A1A8M4L0 A0A2D0T657 A0A3P8PZN6 A0A2Y9REW1 F1QF84 T1J4S4 A0A1S3H6C7 A0A3P9IBC1 H2MCF2 A0A3P9PA00 A0A3P8UWG6 A0A3P8UWH1 V4CNL0 A0A147BMK6 A0A3P9LE65 A0A3B3REW5 A0A341AGN4 A0A2J8WC27 A0A195ED44 A0A2I4C0J8 A0A3Q3BFX4
A0A194RHT7 A0A194PDJ1 A0A3S2LRN5 A0A2H1V1T9 A0A0L7LR60 A0A336KPG1 D6WAH1 A0A087ZQP9 A0A3L8E3Q0 A0A2C9H6G0 A0A1B6IMY3 E0VKZ8 A0A1J1J8M9 J9KBD8 A0A2S2NY28 A0A2H8TL04 A0A2S2QZX7 A0A3B3CL08 A0A1A8CWI6 Q4SZ08 A0A1A8U0B2 A0A1A8MM39 A0A1A8QWS5 A0A1A8FI83 A0A1S3SGI6 A0A146VID1 A0A1A8IGM2 A0A146Q8K5 A0A146PTH2 A0A146VHJ9 A0A1S3MZK1 A0A146ZS10 A0A3P9CVB1 A0A3Q3KLT7 A0A3Q3N444 A0A1A7YQD9 A0A3B1KBM3 A0A1A8M4L0 A0A2D0T657 A0A3P8PZN6 A0A2Y9REW1 F1QF84 T1J4S4 A0A1S3H6C7 A0A3P9IBC1 H2MCF2 A0A3P9PA00 A0A3P8UWG6 A0A3P8UWH1 V4CNL0 A0A147BMK6 A0A3P9LE65 A0A3B3REW5 A0A341AGN4 A0A2J8WC27 A0A195ED44 A0A2I4C0J8 A0A3Q3BFX4
PDB
5LVV
E-value=9.23307e-12,
Score=172
Ontologies
GO
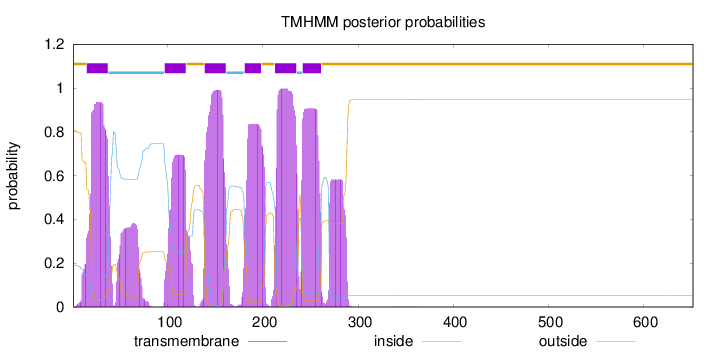
Topology
Length:
652
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
134.103590000001
Exp number, first 60 AAs:
25.26578
Total prob of N-in:
0.19192
POSSIBLE N-term signal
sequence
outside
1 - 14
TMhelix
15 - 37
inside
38 - 96
TMhelix
97 - 119
outside
120 - 138
TMhelix
139 - 161
inside
162 - 180
TMhelix
181 - 198
outside
199 - 212
TMhelix
213 - 235
inside
236 - 241
TMhelix
242 - 261
outside
262 - 652
Population Genetic Test Statistics
Pi
20.554832
Theta
19.084152
Tajima's D
1.340369
CLR
0.894419
CSRT
0.757562121893905
Interpretation
Uncertain
