Gene
KWMTBOMO07576
Pre Gene Modal
BGIBMGA013696
Annotation
PREDICTED:_uncharacterized_protein_LOC106139968_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 2.969
Sequence
CDS
ATGCACGCTCTCCAAATTTTAAGAAGTGGATTACGTTTACCTTTTTTTGTTGATAGATTGAGTATCAGCACCACAGCTAAGAGTTTACGAAAACGATCACCTTGTGACGTCTGCCTTTACCCAACTGACAAGAAACCGTGTTTCGACCTGCGGAATGTATCCATTGAGTTCGGCAACCAATGTCACGCGAAATTAATACGATCGTTCCTCTACACTCATTTCTGGCCTCGGGAGCCCAGTGTCGTTGGTCTGTGGATGTCTTTGGAATGCCCTTACCTTGAAGTTCTGACTGAAAAGTACTCTAATTCAGGAGATCGGTTCTTGGCCTTTGAGCGGATCCAACGGACAGGCGAGCGGAAATTGGCAGGAGTTTGCGTAGCTAATACGGTCAGACCTTGGAAAATCAAGGAATTGGAAGAATGGGCTCACAATACCGACTCAAAACCAGAGAGGCACCGCATGTATTTCTGTGCTCACTGTTTGAAGAGTCCGAACTTATTTGCTAAGTACAACGTTGATTACATCTACGATGTCGAAGTACTGACGACCGCAGCAGAAGTGACAGGTCAGGGACTGGCCACGCTTCTACTTCGCACGGCGCTGGCTCACGCGAATGAGCTGCGTCATCCATTGGTACAAACTGTGGCGGTCAGTCAGTACACATCCAAAATATGCGAAAGAGTCGGTATGCGACGGGAATGGTCTATGAATTATGCCGACTACGTGAACGACGCAGGTCAGCGCGTGTTCTTCCCCCGAAGACCGCACCACACGGTCGCTATTTATGTGAAACATTTTGATCCTAAAAAGGGTGGTGAGGTTCCGTGCAAGCCTCCATATTAA
Protein
MHALQILRSGLRLPFFVDRLSISTTAKSLRKRSPCDVCLYPTDKKPCFDLRNVSIEFGNQCHAKLIRSFLYTHFWPREPSVVGLWMSLECPYLEVLTEKYSNSGDRFLAFERIQRTGERKLAGVCVANTVRPWKIKELEEWAHNTDSKPERHRMYFCAHCLKSPNLFAKYNVDYIYDVEVLTTAAEVTGQGLATLLLRTALAHANELRHPLVQTVAVSQYTSKICERVGMRREWSMNYADYVNDAGQRVFFPRRPHHTVAIYVKHFDPKKGGEVPCKPPY
Summary
Uniprot
H9JVY3
A0A212EMG5
A0A2A4JI98
A0A2H1VQB9
A0A3S2LDI4
A0A0N1I7Q1
+ More
A0A194PDF6 A0A0L7LAI5 A0A1W4X294 B0X0P5 W8BWT5 A0A1Q3G5B6 A0A1J1J6G6 A0A0K8V027 A0A1Y1KEQ6 D2A2Z2 D6WAG9 A0A0A1XMY8 A0A2S2N683 A0A182GD84 A0A034WQ09 Q17C58 A0A1S4F890 A0A1Y1MVH1 D2A0F0 A0A1J0AHX8 N6T6F0 A0A139WJ51 B4KFZ9 A0A034W7A7 A0A034W3K8 R4VGP4 A0A0K8V1D6 A0A023ELE2 A0A2S2P864 A0A1Y1MM63 C4WVB7 A0A034W632 A0A2J7QTX8 R4VE18 A0A1J1I4L5 A0A0A1XGC7 A0A034W625 A0A2P8Z8Y5 A0A0K8W7E8 A0A0A1WHH0 A0A034W7Q8 A0A0A1XTJ4 A0A0K8VZW0 A0A034W8S8 A0A2S2QAQ0 A0A151JXI6 R4V140 W8BHP5 A0A1Q3FKK9 C4WSQ4 A0A0L0CQX2 B4JL23 B4KNV5 A0A0R3P261 B4GTZ1 A0A1B0AG00 Q29FQ9 A0A195CLK4 A0A1B6FHD1 A0A2J7PVW3 A0A1I8PCC7 A0A1L8DNX5 A0A0Q9X7Z8 A0A1L8DBS8 A0A2J7R9K1 R4V906 A0A1L8DCL9 A0A182NV96 A0A1B6G7D1 B4M8C7 A0A1I8PC73 R4UYH5
A0A194PDF6 A0A0L7LAI5 A0A1W4X294 B0X0P5 W8BWT5 A0A1Q3G5B6 A0A1J1J6G6 A0A0K8V027 A0A1Y1KEQ6 D2A2Z2 D6WAG9 A0A0A1XMY8 A0A2S2N683 A0A182GD84 A0A034WQ09 Q17C58 A0A1S4F890 A0A1Y1MVH1 D2A0F0 A0A1J0AHX8 N6T6F0 A0A139WJ51 B4KFZ9 A0A034W7A7 A0A034W3K8 R4VGP4 A0A0K8V1D6 A0A023ELE2 A0A2S2P864 A0A1Y1MM63 C4WVB7 A0A034W632 A0A2J7QTX8 R4VE18 A0A1J1I4L5 A0A0A1XGC7 A0A034W625 A0A2P8Z8Y5 A0A0K8W7E8 A0A0A1WHH0 A0A034W7Q8 A0A0A1XTJ4 A0A0K8VZW0 A0A034W8S8 A0A2S2QAQ0 A0A151JXI6 R4V140 W8BHP5 A0A1Q3FKK9 C4WSQ4 A0A0L0CQX2 B4JL23 B4KNV5 A0A0R3P261 B4GTZ1 A0A1B0AG00 Q29FQ9 A0A195CLK4 A0A1B6FHD1 A0A2J7PVW3 A0A1I8PCC7 A0A1L8DNX5 A0A0Q9X7Z8 A0A1L8DBS8 A0A2J7R9K1 R4V906 A0A1L8DCL9 A0A182NV96 A0A1B6G7D1 B4M8C7 A0A1I8PC73 R4UYH5
Pubmed
EMBL
BABH01036548
AGBW02013845
OWR42667.1
NWSH01001302
PCG71787.1
ODYU01003801
+ More
SOQ43019.1 RSAL01000203 RVE44392.1 KQ460545 KPJ14079.1 KQ459606 KPI91043.1 JTDY01002058 KOB72216.1 DS232242 EDS38287.1 GAMC01000785 JAC05771.1 GFDL01000047 JAV34998.1 CVRI01000067 CRL06473.1 GDHF01020146 GDHF01004355 GDHF01000639 JAI32168.1 JAI47959.1 JAI51675.1 GEZM01086746 JAV58998.1 KQ971338 EFA02243.1 KQ971312 EEZ98630.1 GBXI01002349 JAD11943.1 GGMR01000026 MBY12645.1 JXUM01242248 KQ635428 KXJ62473.1 GAKP01001276 JAC57676.1 CH477311 EAT43933.1 GEZM01019867 JAV89549.1 EFA02503.2 KX812753 APB53910.1 APGK01042001 KB741000 ENN75799.1 KYB28020.1 CH933807 EDW12125.2 GAKP01008780 JAC50172.1 GAKP01008781 JAC50171.1 KC626038 AGM38201.1 GDHF01019904 JAI32410.1 GAPW01003545 JAC10053.1 GGMR01012976 MBY25595.1 GEZM01033272 JAV84297.1 AK341445 BAH71837.1 GAKP01008778 JAC50174.1 NEVH01011192 PNF32023.1 KC626037 AGM38200.1 CVRI01000038 CRK93822.1 GBXI01004322 JAD09970.1 GAKP01008783 JAC50169.1 PYGN01000143 PSN52966.1 GDHF01005312 JAI47002.1 GBXI01016156 JAC98135.1 GAKP01008782 JAC50170.1 GBXI01000139 JAD14153.1 GDHF01007871 JAI44443.1 GAKP01008779 JAC50173.1 GGMS01005626 MBY74829.1 KQ981567 KYN39822.1 KC626025 KC626027 KC626028 AGM38188.1 AGM38190.1 AGM38191.1 GAMC01013784 JAB92771.1 GFDL01006904 JAV28141.1 ABLF02035746 AK340292 BAH70924.1 JRES01000032 KNC34753.1 CH916370 EDW00276.1 CH933808 EDW09000.2 CH379065 KRT07288.1 CH479190 EDW26011.1 EAL31520.2 KQ977622 KYN01362.1 GECZ01020172 JAS49597.1 NEVH01020940 PNF20477.1 GFDF01005963 JAV08121.1 KRG04412.1 GFDF01010294 JAV03790.1 NEVH01006586 PNF37492.1 KC626026 AGM38189.1 GFDF01009863 JAV04221.1 GECZ01011536 JAS58233.1 CH940653 EDW62403.1 KC626033 KC626034 AGM38196.1 AGM38197.1
SOQ43019.1 RSAL01000203 RVE44392.1 KQ460545 KPJ14079.1 KQ459606 KPI91043.1 JTDY01002058 KOB72216.1 DS232242 EDS38287.1 GAMC01000785 JAC05771.1 GFDL01000047 JAV34998.1 CVRI01000067 CRL06473.1 GDHF01020146 GDHF01004355 GDHF01000639 JAI32168.1 JAI47959.1 JAI51675.1 GEZM01086746 JAV58998.1 KQ971338 EFA02243.1 KQ971312 EEZ98630.1 GBXI01002349 JAD11943.1 GGMR01000026 MBY12645.1 JXUM01242248 KQ635428 KXJ62473.1 GAKP01001276 JAC57676.1 CH477311 EAT43933.1 GEZM01019867 JAV89549.1 EFA02503.2 KX812753 APB53910.1 APGK01042001 KB741000 ENN75799.1 KYB28020.1 CH933807 EDW12125.2 GAKP01008780 JAC50172.1 GAKP01008781 JAC50171.1 KC626038 AGM38201.1 GDHF01019904 JAI32410.1 GAPW01003545 JAC10053.1 GGMR01012976 MBY25595.1 GEZM01033272 JAV84297.1 AK341445 BAH71837.1 GAKP01008778 JAC50174.1 NEVH01011192 PNF32023.1 KC626037 AGM38200.1 CVRI01000038 CRK93822.1 GBXI01004322 JAD09970.1 GAKP01008783 JAC50169.1 PYGN01000143 PSN52966.1 GDHF01005312 JAI47002.1 GBXI01016156 JAC98135.1 GAKP01008782 JAC50170.1 GBXI01000139 JAD14153.1 GDHF01007871 JAI44443.1 GAKP01008779 JAC50173.1 GGMS01005626 MBY74829.1 KQ981567 KYN39822.1 KC626025 KC626027 KC626028 AGM38188.1 AGM38190.1 AGM38191.1 GAMC01013784 JAB92771.1 GFDL01006904 JAV28141.1 ABLF02035746 AK340292 BAH70924.1 JRES01000032 KNC34753.1 CH916370 EDW00276.1 CH933808 EDW09000.2 CH379065 KRT07288.1 CH479190 EDW26011.1 EAL31520.2 KQ977622 KYN01362.1 GECZ01020172 JAS49597.1 NEVH01020940 PNF20477.1 GFDF01005963 JAV08121.1 KRG04412.1 GFDF01010294 JAV03790.1 NEVH01006586 PNF37492.1 KC626026 AGM38189.1 GFDF01009863 JAV04221.1 GECZ01011536 JAS58233.1 CH940653 EDW62403.1 KC626033 KC626034 AGM38196.1 AGM38197.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000283053
UP000053240
UP000053268
+ More
UP000037510 UP000192223 UP000002320 UP000183832 UP000007266 UP000069940 UP000249989 UP000008820 UP000019118 UP000009192 UP000235965 UP000245037 UP000078541 UP000007819 UP000037069 UP000001070 UP000001819 UP000008744 UP000092445 UP000078542 UP000095300 UP000075884 UP000008792
UP000037510 UP000192223 UP000002320 UP000183832 UP000007266 UP000069940 UP000249989 UP000008820 UP000019118 UP000009192 UP000235965 UP000245037 UP000078541 UP000007819 UP000037069 UP000001070 UP000001819 UP000008744 UP000092445 UP000078542 UP000095300 UP000075884 UP000008792
PRIDE
Interpro
SUPFAM
SSF55729
SSF55729
ProteinModelPortal
H9JVY3
A0A212EMG5
A0A2A4JI98
A0A2H1VQB9
A0A3S2LDI4
A0A0N1I7Q1
+ More
A0A194PDF6 A0A0L7LAI5 A0A1W4X294 B0X0P5 W8BWT5 A0A1Q3G5B6 A0A1J1J6G6 A0A0K8V027 A0A1Y1KEQ6 D2A2Z2 D6WAG9 A0A0A1XMY8 A0A2S2N683 A0A182GD84 A0A034WQ09 Q17C58 A0A1S4F890 A0A1Y1MVH1 D2A0F0 A0A1J0AHX8 N6T6F0 A0A139WJ51 B4KFZ9 A0A034W7A7 A0A034W3K8 R4VGP4 A0A0K8V1D6 A0A023ELE2 A0A2S2P864 A0A1Y1MM63 C4WVB7 A0A034W632 A0A2J7QTX8 R4VE18 A0A1J1I4L5 A0A0A1XGC7 A0A034W625 A0A2P8Z8Y5 A0A0K8W7E8 A0A0A1WHH0 A0A034W7Q8 A0A0A1XTJ4 A0A0K8VZW0 A0A034W8S8 A0A2S2QAQ0 A0A151JXI6 R4V140 W8BHP5 A0A1Q3FKK9 C4WSQ4 A0A0L0CQX2 B4JL23 B4KNV5 A0A0R3P261 B4GTZ1 A0A1B0AG00 Q29FQ9 A0A195CLK4 A0A1B6FHD1 A0A2J7PVW3 A0A1I8PCC7 A0A1L8DNX5 A0A0Q9X7Z8 A0A1L8DBS8 A0A2J7R9K1 R4V906 A0A1L8DCL9 A0A182NV96 A0A1B6G7D1 B4M8C7 A0A1I8PC73 R4UYH5
A0A194PDF6 A0A0L7LAI5 A0A1W4X294 B0X0P5 W8BWT5 A0A1Q3G5B6 A0A1J1J6G6 A0A0K8V027 A0A1Y1KEQ6 D2A2Z2 D6WAG9 A0A0A1XMY8 A0A2S2N683 A0A182GD84 A0A034WQ09 Q17C58 A0A1S4F890 A0A1Y1MVH1 D2A0F0 A0A1J0AHX8 N6T6F0 A0A139WJ51 B4KFZ9 A0A034W7A7 A0A034W3K8 R4VGP4 A0A0K8V1D6 A0A023ELE2 A0A2S2P864 A0A1Y1MM63 C4WVB7 A0A034W632 A0A2J7QTX8 R4VE18 A0A1J1I4L5 A0A0A1XGC7 A0A034W625 A0A2P8Z8Y5 A0A0K8W7E8 A0A0A1WHH0 A0A034W7Q8 A0A0A1XTJ4 A0A0K8VZW0 A0A034W8S8 A0A2S2QAQ0 A0A151JXI6 R4V140 W8BHP5 A0A1Q3FKK9 C4WSQ4 A0A0L0CQX2 B4JL23 B4KNV5 A0A0R3P261 B4GTZ1 A0A1B0AG00 Q29FQ9 A0A195CLK4 A0A1B6FHD1 A0A2J7PVW3 A0A1I8PCC7 A0A1L8DNX5 A0A0Q9X7Z8 A0A1L8DBS8 A0A2J7R9K1 R4V906 A0A1L8DCL9 A0A182NV96 A0A1B6G7D1 B4M8C7 A0A1I8PC73 R4UYH5
PDB
4FD6
E-value=3.51615e-06,
Score=119
Ontologies
GO
Topology
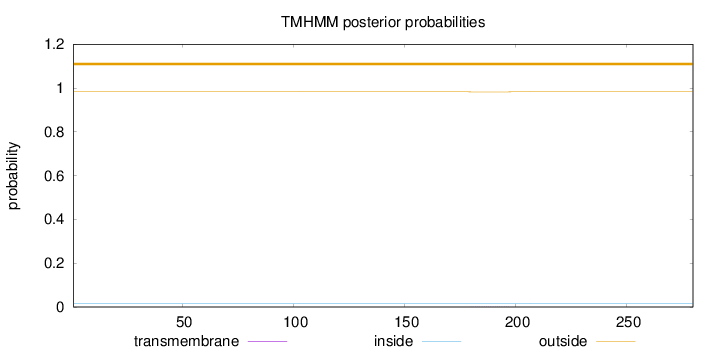
Length:
280
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00502
Exp number, first 60 AAs:
0.00038
Total prob of N-in:
0.01741
outside
1 - 280
Population Genetic Test Statistics
Pi
172.76708
Theta
146.011586
Tajima's D
0.572698
CLR
0
CSRT
0.539623018849058
Interpretation
Uncertain
