Gene
KWMTBOMO07572
Pre Gene Modal
BGIBMGA013729
Annotation
PREDICTED:_facilitated_trehalose_transporter_Tret1-like_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.366
Sequence
CDS
ATGGGATCTCAAAATTGGGACACATACATTCGTCGTGTAAAACAATTCATGGAACTCAACGACATTGAACAGAGACTTCACGTTGCAACGCTCGTGACCTTAGTTGGGTCTGAGTGCTATGACTTAATGTGTGACTTGTGTGCTCCGGACTTACCAGAAGAAAAGGATTTCGACATACTCGTAAAGTTAGTCAAAGATCATCTAGAACCTGAAAGATCGGAAATAGCAGAACGTCATATCTTCAGGCAAAGAAAGCAGCAAAATAACGAAAGCATTCGTTCCTACCTACAAAGTTTGAAACATTTGGCTAAAACTTGCAATTTCGGCATCACATTAGAAATTAATCTACGCGATCAATTCGTATCAGGATTATATAGTGAGGAGATGCGATCAAGGCTGTTTGCTGAAAAAGATATAGATTACAAGCGCGCTGTTGAGTTGGCATCAGCACTGGAAGCAGCGGATCGACATGCGATGGCGGCTGGGGGCACTGGGGGCAGCTCACAGGCTGCGTCCGAGAGCGATGGCATGCATCGGGTCGGCGGAGCGCGCGCGGAGGCTCGGTGCGCCTGCAGACGCTGCGGCAAGCTGGGCCATGCGGAGGGTCGGTGTCGGTACAAGCACTACACCTGTGACTCATGTGGCGAGAAGGGACACCTAAAATCAATGTGTCGGAACCGTAAAGGTAACCAAAAGATTGAAAAAACCCGAGAAAGCTTTCCACTCAGTTTTAAAGAAATTAAAAAGGAAGTTACGAAGGATCCTTTACTAAATAAAATTTACGGTTACGTCATGTTTGGCTGGCCCTCTGTCAGTAAAGAACAAGCAGAAAAACCCTATTTTAACAGAAGAGAATCCATACATATTGATCACGGTTGTCTTGTATGGGGTTACAGAATAATAATTCCAACTTCATTGCGAGAGACTGTGTTACAGGAGTTACATGAGGGACATCCGGGTATTGTTAAGATGAAACAATTAAGTCGGAACTATGTATGGTGGGAGACAGTCGATGCGGACATTGAACGTGTATGTGCTCAGTGCCAGGCCTGTCGGTCTCAGCGCGCTGCACCACCATCGGTGCCGCTGCACTCATGGCCATGGCCGGAAGAGCCGTGGGCGCGCCTTAATCTTGATTTCTTGGGACCGTTTAATAACAAATATTACTTAATTATAGTTGACGCTCATTCAAAATGGATTGAGGTGGAAAAATTGAGCGGTACATCAGCCACGGCAGTAATCGCTTGTCTCAGACGATTGTTTGCTCGATTTGGCTTAGCAAAAAGGGTAGTAACTGATAACGGACCACCTTTCTCGTCGGTCGAATTTAGATCATATTTAAGTAAAAACGGAATAAAACATTTACCGGTGGCTCCGTATCATCCATCTAGCAATGGTGCAGCGGAAAACGCCGTAAAAACCATAAAAAGGGTGTTGAAAAAGGCTCTAATTGAAAAGGAAGACGACAATACCGCCTTGACAAAATTTCTCTTTATGTACAGAAATTCTGAGCATAGCACTACAGGGCGCGAACCAGCAGTTGCTTTGTTCGGACGTCGCCTGCGTGGAAGACTGGATCTGTTGCGTCCCAATACCAGCTCCATAGTACGAGAAGCACAGCTGGCAAGCGAAAGCTATAATGCAGGCACAGCCGGATATAGGGAGGCTGCAGAAGGAGACTCGGTCCTATTGCAAGATTTCACACAGGGAAAAAAGAAATGGACCACAGGAACAATAAAAAATCGCTCAGAGGACATGTCACAAACAAGTGAAGAATCTACGAAAAAAGGACACACGTACCTGCAGTGGACTTTCTCTATAATATCAAACATAGCCTACTTGAATTACGGGTTGCAAAATGGTTGGATTTCACCGTTCGCAAAAAAACTACAGTCAGAAGACTCACCGACCGGGCGACCGTTGACCAGCACTGAGTTTTATTGGGTAGCTAGTGTATCATCACTGGCTGCAATGTTTGGTGTGTCAGCATTCGCGGCCTTAGCTGATGGACTAGGCAGAAAAATCGGGGTCGTCGCCGTGGCTGCGGTGCAGGCTCTCTGCTGGATCATAAAACTGTGTCCTTCAACAACAGTAACTCTGATAATTGCGAGAATCTGTAGCGGCCTGGGAGCTGGTGGAGCTTTCGCGATAGTCCCAATGTATATAAAAGAAATTAGCCAGAACAATATTAGAGGAACCCTCGGAGTCATCATGACAATGTCTCAGAATTCTGGATATTTGATAATGTATTCGATGGGAGCATATATGGAGTATTATACGGTACTATGGATAGCGGCTGTTCCGCCGATACTGAATCTGTTGCTGATGCTATTCGCGCCAGAATCTCCTGCGTTCCTATTGAAAAAAGGAAAAGTCAGTGAAGCTACTGAAGTGATAGCAGGCTTAAGAGGCTTAAATATTGATGACAATGTAGTTTTAAATGAGATTAATGAAATAAGAAATGAAGACGATTTTTACAAGTCTATACCCAATATTTCGTTTTTTGGAATCTTTAAACAGCCAGTATGGCGTCGTGGATTTTTAATAATGTTCGTAATAATTACCACCCATGGTCTAAACGGCGTATTTACTATAGTAACATATGCTTCAACGATCCTGCGAGCCTCTGGAATAACATTAAATCCTGAACTGCAGTTATTAAGTATACCAGCCTTTATGATAATATCTTCAGCTCTGTCCATAATGCTTGTGGAAAGAATGGGAAGAAAGTTCCTTCTAGCAGGTACATATATAGTATCAGCATTCTCATGTGGAACATTAGCTACGACGTTACTAATGCAGGAAAATGGATTCGCTACACCAGGCTGGTTGCCAATTGTAGCTATTGTAGCTACGGTAAGCTGCTATGCGGCTGGAATTTTGCCTCTTCCTTTCGTTATTATGTCTGAGATGTTTAACTTCCAGATCAGAGCAAAAGTGATGAGTTGCATAATAACCACAGGCTGGTTAATGTCATTCGTCCAAGCAGCATCTTATGGACCAATCACAGAAATTTTAGGCTCCTCCGCAGCCTTTTACTTTTTCGCTTTCGTTAATTTTTATGGTGCAGTCACTGCTCTGTTCTTTTTACCCGAAACAAAAGGTAAAAGTGTGGAAGAAATTGAATTTGAATTAAAAGCAAAGTAG
Protein
MGSQNWDTYIRRVKQFMELNDIEQRLHVATLVTLVGSECYDLMCDLCAPDLPEEKDFDILVKLVKDHLEPERSEIAERHIFRQRKQQNNESIRSYLQSLKHLAKTCNFGITLEINLRDQFVSGLYSEEMRSRLFAEKDIDYKRAVELASALEAADRHAMAAGGTGGSSQAASESDGMHRVGGARAEARCACRRCGKLGHAEGRCRYKHYTCDSCGEKGHLKSMCRNRKGNQKIEKTRESFPLSFKEIKKEVTKDPLLNKIYGYVMFGWPSVSKEQAEKPYFNRRESIHIDHGCLVWGYRIIIPTSLRETVLQELHEGHPGIVKMKQLSRNYVWWETVDADIERVCAQCQACRSQRAAPPSVPLHSWPWPEEPWARLNLDFLGPFNNKYYLIIVDAHSKWIEVEKLSGTSATAVIACLRRLFARFGLAKRVVTDNGPPFSSVEFRSYLSKNGIKHLPVAPYHPSSNGAAENAVKTIKRVLKKALIEKEDDNTALTKFLFMYRNSEHSTTGREPAVALFGRRLRGRLDLLRPNTSSIVREAQLASESYNAGTAGYREAAEGDSVLLQDFTQGKKKWTTGTIKNRSEDMSQTSEESTKKGHTYLQWTFSIISNIAYLNYGLQNGWISPFAKKLQSEDSPTGRPLTSTEFYWVASVSSLAAMFGVSAFAALADGLGRKIGVVAVAAVQALCWIIKLCPSTTVTLIIARICSGLGAGGAFAIVPMYIKEISQNNIRGTLGVIMTMSQNSGYLIMYSMGAYMEYYTVLWIAAVPPILNLLLMLFAPESPAFLLKKGKVSEATEVIAGLRGLNIDDNVVLNEINEIRNEDDFYKSIPNISFFGIFKQPVWRRGFLIMFVIITTHGLNGVFTIVTYASTILRASGITLNPELQLLSIPAFMIISSALSIMLVERMGRKFLLAGTYIVSAFSCGTLATTLLMQENGFATPGWLPIVAIVATVSCYAAGILPLPFVIMSEMFNFQIRAKVMSCIITTGWLMSFVQAASYGPITEILGSSAAFYFFAFVNFYGAVTALFFLPETKGKSVEEIEFELKAK
Summary
Uniprot
ProteinModelPortal
PDB
4LDS
E-value=4.06102e-29,
Score=323
Ontologies
GO
Topology
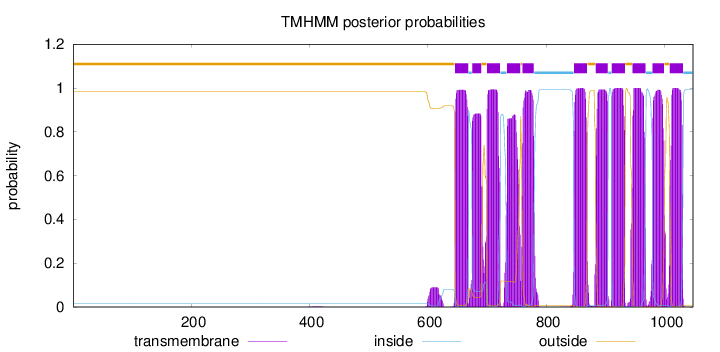
Length:
1048
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
235.19904
Exp number, first 60 AAs:
0.00127
Total prob of N-in:
0.01600
outside
1 - 645
TMhelix
646 - 668
inside
669 - 674
TMhelix
675 - 690
outside
691 - 699
TMhelix
700 - 722
inside
723 - 733
TMhelix
734 - 756
outside
757 - 759
TMhelix
760 - 779
inside
780 - 846
TMhelix
847 - 869
outside
870 - 883
TMhelix
884 - 904
inside
905 - 910
TMhelix
911 - 933
outside
934 - 945
TMhelix
946 - 968
inside
969 - 979
TMhelix
980 - 999
outside
1000 - 1008
TMhelix
1009 - 1031
inside
1032 - 1048
Population Genetic Test Statistics
Pi
230.631401
Theta
191.577113
Tajima's D
0.455796
CLR
0
CSRT
0.500074996250187
Interpretation
Uncertain
