Gene
KWMTBOMO07570
Pre Gene Modal
BGIBMGA013730
Annotation
PREDICTED:_fat-like_cadherin-related_tumor_suppressor_homolog_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.204 Nuclear Reliability : 1.175
Sequence
CDS
ATGTCAGATGTGGCGCTCGCATGGCTGGTGGTGGCGGCGGCAGTCTCATGGTGCGCGGGCGCCGGCCCCCCCGCCTTCCGCTTCCACCATGCGCTCTACAACCTGACCGTGCCCGAAAACAGTCCGCCTCGCACGTACGCCGCGCCGCCGCCCGCCGCCGCGCCGCTCGGGGTGCGTCTGCCCGACCCGCACGCGCGCGTCCGGTTTCGCATCAGACACGGAGATCGAGACAAATTCTTCAAGGCCGAAGAGCGCACCGTCGGTGACTTCTGCTTCCTGTCGATTAGGACGCGCACCGGACAAACCGACGTGCTCAACCGCGAGCGCCGCGACTCTTATCAGCTGGACGTGCGCGCCACCGCGCTGCTCCCCGGCGGCCGCCAACTCGAGGCAGACACCATCGTCTTGATCCAGATATCGGATGAAAACGACCTTAGTCCGCTGTTCTACCCGACGGAATACGAAGCATCAGTGCCCGAGGACGCTCCGCCTCATTCTAGTGTAATCCGAGTGTCTGCGGAGGATGCCGACCTCGGCCTCAATGGAGAAATCTACTACAGCCTGGCCGACTCGATCGAACGATTCGCAATACACCTCACCACCGGCGTGATAACTATAACAAGACCTTTATCTGCAGCTGAAAAATCTAAGTACGAGTTGACGGTTCTGGCGAGGGACCGCGCGTCTTTGCTCGCGCACGGAGAGGAAGCACCCGCGGCCCGAGCCTCGGTAACGGTGCGCGTCAGACGCGTCAACCTGCACGCTCCAGAGCTTCGGGTTCGGAGACTGCCAGAGATAGTGGAAAATTCGACAACAGAAATATATGCTATAGTGGAAGTGAGCGATCGCGACGTAGGCGTGAACGGCCGCATCGCCGGTCTGGACATAGTGGACGGCGACCCAGACGGACACTTCCGAGTGCGTCCGTCCAGCCAGCCAGGCGAGTTCGACGTCGTAGTGCATTCATTACTCGATCGCGAAACTGCACCCCTCGGATATAACTTAACCTTGCGAGCGATGGATGCGGGCGTGCCGCCGAGAGCCTCGTACGTGACCGTGCCCGTGACGCTCATAGACGCTAACGACAACGCGCCGGTGTTCAGCCGAGAAGTGTACGAGGCGACCGTGTCTGAGACGGCACCACCCAACACGCCCGTCATCCGCCTCAAGGTCACCGACAGGGACGAGGGCCGGAACGCCCGCGTCTACATCGAGGTGGTCGGAGGGAACGAGGGCGGAGAATTCCGCGTCAACCCGGAGACTGGCGTGCTCTACACTGCGGTGCCTCTCGACGCCGAGGAGAAAGCCTTCTATACCCTCACCGTCTCGGCAATCGATCAGGGAAACGCGGGCACGAGAAAACAATCATCCGCTAAAATCAAAATATCCGTTGCCGACGCTAACGACAACGATCCAGTCTTTGAACAAAAGGAGATGTCTGTGAAAGTCCCGGAGAACGGACAAAGCGGCGGCGTGGTGGCGCGCGTCGCGGCGCGGGACGCGGACTCGGGGGATAACGCCTACGTCTCGTACAGCATCGCCAACATACAACCCGTCCCGTTCGACGTGGACCACTTCTCCGGTGCGGTGCGGACCACGCGACTGCTGGACTACGAGAGCATGCGACGCGAGTACGTGCTGCGAATACGAGCCAGTGACTGGGGCCTCCCGTACCGACGCCAGGCCGAAATGAGACTCGTCGTGCAACTCAACGACGTCAACGACAACAGACCGCAGTTTGAGAGGGTGGACTGCGTGGGCTACGTCCCGCGCCGCGTCGCCATCGGCTCGGAAATAGTGACGCTCTCGGCTATTGACTTTGATGATGGCGACGTGGTTTCTTATCGAATCGTCGGCGGCAATGATGATAATTGCTTCTCGTTGGATTCGAGCACGGGCGTCGTGAGTCTCGCCTGCGATCTGAACGACATCCGAGCCGACTCCCGCGTGCTGAACGTGAGCGCGACAGACGGGACACACTTCGCCGATGCAACGTCGTTGGTTTTACACCTAGTGTCGGGAGGCGGCGAGAGCGGGGCCCTCGAGTGTCGCGACACCGGCGTAGCGCGTCGCCTCACCGAACTGCTGGCGGCCGCTGAACGCAGTAATGCACCGCTGGATTTTGCAGATGAATTTCCCTTAGCACCGTCTCGTTATGGTGAAAATTTACATTCCCCCGAATTCGTCAATTTCCCCATTCAAGTCAAAGTAAACGAATCTGTTGCATTAGGAACATCGTTAGTGCGGCTGCAAGCTCGCGACCACGACCTCGGCTACAATGGACTCATCACGTACGGTATATCCGGCGGAGACGCCGATTCGGCGTTCAGAATCGACCCTGAAAACGGTGATCTGCAAGTGATCGGTTACCTGGACCGGGAACGCGAAAGCGAGTATTATTTGAATGTGACAGCATACGATCTCGGGCGACCGCAGCGCTCCACGTCACGCATGCTGCCCGTCATCGTCCTCGATGTGAACGACAATCGCCCGCGGCTCCAGAAAACTTTGGCGAGCTTCCGAGTGACGGAGAACGCTTTGAACGGAACCGAAGTCTTCCGAGCGAACGCCACCGACCGCGACGCCGGCGAGTTCGGGAGAGTGACGTACAGCCTGAGCGGCGCCACCGAGGGGGAGTTCTGCATCGAGCGCGAGAGCGGCGCGCTGACAGTGTGCGCGCCGCTGGACCGCGAGCGCCGCGCGCTGTACGAGCTCACGGTGCGCGCGGCCGACGGCGGCGGGCTGCACGCGGAGGCGCTCGTGCGGGTCGCGGTGGACGACGTCAACGACAACGCGCCGCGCTTCGGGCTCGGCTCGTACGCAGCCAGGGTGCGGGAGGACGTGCCCGCGGGGACGCTGGTGGCCGTGCTCGATGCCTTCGACCCGGACACCGCCGCGGGCGGACAGATCACTTACTCCCTCTCTGACCAAGAAGAAGAAGCCGTGTTCACGATCGATCCCGTCTCGGGAACGTTGCGCACCGCGCGCTCGCTCGACTTCGAACAACGACAGGTGTACGGCGTGACGGTGCGGGCCACGGACGGCGGGCGGCCGGCGCTGTGGAGCGAGGCCACGGTCATCGTCGAGGTGATCGACGTGGACGAAAACACGCATGCGCCGGCGTGGGACGCGCACGACGTGCTGGCGGGCTCCGTGCGCGAGGACGCGCCGCGGGGGACGCGCGTGCTGGCGGCGCCGGCCGCGGACGCCGACCCGCCAGGGAGGGACTCCCGGCTCGCCTACTACATAGTGGCCGGCTCCGGGATGGCGCACTTCTCCGTCGATGACGCTGGTATCATCAGAACGTTGACGCCCCTTGATAGAGAATCCGTGCCTCACTATTGGCTGACGTTGTGCGCACAAGATCACGGACTCATACCTAAACATTCGTGCATTCAGGTGTACATAGAAGTGGAAGACGTGAACGACATGGTCCCGTGGCCGGCGCGCGCGCGGTACGAGGCCGCGGTCCCCGAGCACTGCGCCGCGGGGACCAGCGTCACGCGCGTCACGGCGCTCGACGCGGACGACGCGCCGCACTCCAACCGCACGGCCGTCACGTACTCCATCGTCGCAGGAAACCCCGACGGACTGTTCTCGATCGACGAGCGAACAGGTGAGATCGTGACGACGGGACGAAGCCTGGACCGAGAGTCGGCAGCAACCCACGCGCTAGAGGTGCAGTGCTCGGACGGAGAGCTCAGCAGCACCACGCGGGTGCACGTGCGCCTCCTCGACCTCAACGACCACAGCCCCGTCTTCACTCAGCGCCTGTACGACGTGCATGTGCCGGTGCCGCCCCGACCTGACCTCCTCGCGCAGAGTGACGCTGCCGAAGGGACGACCGACGAGAGCGCCGAGTGGGACACCGACGAAGACGAGGAAGCCGAGGGCGCGCGGTCCAACTGGGAGCAGTGGCCGGACCCGCCCGCCGACGGCGTGCACGTGGCCACGGTGATGGCGTTCGATCCGGACGAGGGCGCGAACGGCACGGTGCGGTACTGGAGCCGGGCCCGCGGCGCCGCGCGCGGCCTGCTGCGCGTCTGCGCACGCACCGGCCGCCTGCTCGCCGCGCCGCGCCTGCCGCTCGCGCCCGCACACGCGCACGACGTCACGGTGCGCGCTTGCGACGGGGGCGCGCGGCCGCGATGTGCAGTGGCCCGGGTGAGCGTGCGCGGTGTTGCGGCACGGGAAGGGGGCGACCCCCCGCGTCTGTCCCCCACCCCTCCTCTGCAAGTGGCCGAGCTCGACGCGCCCGGCTTCCTGCTCGCCGTCCTTCAGGCGACCGATCCGGACTCCGATCCTCTGTACTATGCTATCGTAGAAGGCGACCCCCGCAACGAATTCTTCATCGGGCGCGAGGACGGCACGCTGGTGCTGGCGCGCCGGCTGCTCTGGGAGCGACAGGCGTTTTATTCCCTGAACGTGTCCGTCACCGACGGAACCCACGTCGTGTACTCTTTGGTCAACATAACCGTCATCAACGATGCGAACGAAGACGGCGTGTCGTTCTCGCGTGAAGAGTACGTGGTGGAGACGTCGGAGGGCGCGCGCGTGGGCGAGGCGCTGGTGGTGGTGGGCGCGGCGGTGGGGCGCGGGGCGGCGGGGGGCGCGGCGCGGCTGCTGTACGGGCTGCACGCCGCGCGCGCCCCCGCCTCGCTGCTGCTGTTCCGCCTCAACGAGCTCACCGGCGTGCTCGAACTCGCTCAGCCGCTCGACAGGGAGAGCGCAAGCGTCCACGAGCTCACCGTGTGGGCCCGCGACCAGGCGCCGCGCGCGTCTGTCGCCTTCGCGCGCGTGCGCATCCACGTGCACGACGCGGACGAGCACGCGCCCGTGTGGGGCCGGCGGCTGGCGGAGGCGCGGGTGGCGCGCACCACGGCGCCCGGCACGCTGGTGGCGGCGCTGCGGGCCGCCGACGCCGACGCGGGTGACGCGGCGCGCATCGTCTACTCGCTCGTGGGCGGAGACGCCGCCGGACTGTTCGAGGTGGACGCGACGCTGGGCGACGTGACGCTGAGTCGCGCGCTGCCCGCCCGCGGCCCGCGCGACTACACGCTCAACGTGCGCGCGTCCAACCCGCCGCCCTCCACGCGCTTCTCGACGCTACCGCTCCACGTCACCGTGGTGGACCCGGACGACGCCCCGCCGCGGTTCGTTTTGGAAGAAATATCTTGTGAAGTATACGAGAACGAGCCGGCGGGCGGCACGCTGGTGGCCGCGGAGGCTCGCAGCGGCACGGCGCTCTGGTACACGCTGGAGGGCGGCGCCGGCCTGTTCCGCATCAACCCCGCCGCCGGCCTCATCGCCGCCGTGGCACCCCTGGACTACGAGGCCGCCGCGCTGTACAACCTGACCGTGACCGCGCTCAGCATGGGCGGGGGCTCGGCGAGCGCGCGCGTGACGGTGCACGTGCTGGACCGCAACGAGTTCCCGCCGCGGCTGCTGCGGCGCGAGTACCGCGGGCGCGCGTCGGAGGCGGCGGCGGCGGGCGCGCTGGTGGCGGACGGCGAGGGCGCGCCGCTGGTGCTGCTCACGGAGGACGCGGACTCCGCCGCCAACCGACAGCGCTCCTTCGAGATCGTGGAGCCGGCGGCCGCCGCGCTGTTCCGCGTGGACCCCACCACGGGCGCGCTGCGCCTGGCCGCCGCGCTCGACTACGAGACGGCAGCCGTCCACACCTTCACCGTCAAGGTGCTGGACGCGGGCGAGCCCCGGCTGCCGTCCGACAGCACGGCGACGGTCACCGTCGACGTGATCGACGTCAACGACTGCCCACCCGAGTTCACCGAGTCCACGTACTCGACGACGGTCCTCCTGCCCACCTCGAGCGGCGTCACGGTGCTGGCGCTGCAGGCGCACGACCGCGACCTGCCGCCCGGCGCCGCGCTGCACTACGACATCATCGAGGGCGACGCGGCCGGCGCCTTCGCGCTGTCCGAGTCGGGCGTGCTGTCCGTGTCCCGGCCGGACGCGCTGTCCGACTCGCACCGGCTGCGCGTGCGCGTGTCGGACGGCCGGTACGCGGCCACGGCGCGCGTAGACGTACAGATCCGAGAGGCCGACAACTCCGGTCTCGCCTTTCAAAAATCCGATTACTACGGCACAGTCGTCGAGAACTCGACGAAGCCGGCGAACGTCGCCGTGCTCAACGTGCTCGGCGCGGCGCTCAACGAACACGTGCAGTTCGTCATCCTCAACCCCGTCGACGGCTTCGAAGTGAGTTTTTATTCCTTAGATGGCTGA
Protein
MSDVALAWLVVAAAVSWCAGAGPPAFRFHHALYNLTVPENSPPRTYAAPPPAAAPLGVRLPDPHARVRFRIRHGDRDKFFKAEERTVGDFCFLSIRTRTGQTDVLNRERRDSYQLDVRATALLPGGRQLEADTIVLIQISDENDLSPLFYPTEYEASVPEDAPPHSSVIRVSAEDADLGLNGEIYYSLADSIERFAIHLTTGVITITRPLSAAEKSKYELTVLARDRASLLAHGEEAPAARASVTVRVRRVNLHAPELRVRRLPEIVENSTTEIYAIVEVSDRDVGVNGRIAGLDIVDGDPDGHFRVRPSSQPGEFDVVVHSLLDRETAPLGYNLTLRAMDAGVPPRASYVTVPVTLIDANDNAPVFSREVYEATVSETAPPNTPVIRLKVTDRDEGRNARVYIEVVGGNEGGEFRVNPETGVLYTAVPLDAEEKAFYTLTVSAIDQGNAGTRKQSSAKIKISVADANDNDPVFEQKEMSVKVPENGQSGGVVARVAARDADSGDNAYVSYSIANIQPVPFDVDHFSGAVRTTRLLDYESMRREYVLRIRASDWGLPYRRQAEMRLVVQLNDVNDNRPQFERVDCVGYVPRRVAIGSEIVTLSAIDFDDGDVVSYRIVGGNDDNCFSLDSSTGVVSLACDLNDIRADSRVLNVSATDGTHFADATSLVLHLVSGGGESGALECRDTGVARRLTELLAAAERSNAPLDFADEFPLAPSRYGENLHSPEFVNFPIQVKVNESVALGTSLVRLQARDHDLGYNGLITYGISGGDADSAFRIDPENGDLQVIGYLDRERESEYYLNVTAYDLGRPQRSTSRMLPVIVLDVNDNRPRLQKTLASFRVTENALNGTEVFRANATDRDAGEFGRVTYSLSGATEGEFCIERESGALTVCAPLDRERRALYELTVRAADGGGLHAEALVRVAVDDVNDNAPRFGLGSYAARVREDVPAGTLVAVLDAFDPDTAAGGQITYSLSDQEEEAVFTIDPVSGTLRTARSLDFEQRQVYGVTVRATDGGRPALWSEATVIVEVIDVDENTHAPAWDAHDVLAGSVREDAPRGTRVLAAPAADADPPGRDSRLAYYIVAGSGMAHFSVDDAGIIRTLTPLDRESVPHYWLTLCAQDHGLIPKHSCIQVYIEVEDVNDMVPWPARARYEAAVPEHCAAGTSVTRVTALDADDAPHSNRTAVTYSIVAGNPDGLFSIDERTGEIVTTGRSLDRESAATHALEVQCSDGELSSTTRVHVRLLDLNDHSPVFTQRLYDVHVPVPPRPDLLAQSDAAEGTTDESAEWDTDEDEEAEGARSNWEQWPDPPADGVHVATVMAFDPDEGANGTVRYWSRARGAARGLLRVCARTGRLLAAPRLPLAPAHAHDVTVRACDGGARPRCAVARVSVRGVAAREGGDPPRLSPTPPLQVAELDAPGFLLAVLQATDPDSDPLYYAIVEGDPRNEFFIGREDGTLVLARRLLWERQAFYSLNVSVTDGTHVVYSLVNITVINDANEDGVSFSREEYVVETSEGARVGEALVVVGAAVGRGAAGGAARLLYGLHAARAPASLLLFRLNELTGVLELAQPLDRESASVHELTVWARDQAPRASVAFARVRIHVHDADEHAPVWGRRLAEARVARTTAPGTLVAALRAADADAGDAARIVYSLVGGDAAGLFEVDATLGDVTLSRALPARGPRDYTLNVRASNPPPSTRFSTLPLHVTVVDPDDAPPRFVLEEISCEVYENEPAGGTLVAAEARSGTALWYTLEGGAGLFRINPAAGLIAAVAPLDYEAAALYNLTVTALSMGGGSASARVTVHVLDRNEFPPRLLRREYRGRASEAAAAGALVADGEGAPLVLLTEDADSAANRQRSFEIVEPAAAALFRVDPTTGALRLAAALDYETAAVHTFTVKVLDAGEPRLPSDSTATVTVDVIDVNDCPPEFTESTYSTTVLLPTSSGVTVLALQAHDRDLPPGAALHYDIIEGDAAGAFALSESGVLSVSRPDALSDSHRLRVRVSDGRYAATARVDVQIREADNSGLAFQKSDYYGTVVENSTKPANVAVLNVLGAALNEHVQFVILNPVDGFEVSFYSLDG
Summary
Description
Cadherins are calcium-dependent cell adhesion proteins.
Uniprot
H9JW17
A0A212ER96
A0A139WMQ2
A0A139WN77
A0A0N8ES92
A0A1Y1MVG4
+ More
A0A182LW70 A0A1Y1MVJ3 A0A1I8JVT2 A0A2M4CS75 A0A336KKB1 A0A2L1IQ90 K7J800 E0VDI8 X1X3C6 A0A2H8TXV2 A0A2K8JM25 A0A224X4K1 A0A2M4AFQ1 A0A194PCL9 A0A1J1HSS9 A0A0P5TSL7 A0A0P5BYN3 A0A0P5Y7P4 A0A0P6FPD2 A0A0P5PDR7 A0A0P6ALW4 A0A0N8A6F3 A0A0P5C0D4 A0A0N8BRM6 A0A0P5RZV7 A0A0P6DWR4 A0A0P5NJI0 A0A0P5CZJ4 A0A0P6FWK1 A0A0P6DMB0 A0A0N8BMQ8 A0A0P5CUN3 A0A0P5S461 A0A0P6HTE5 A0A0P5WR69 A0A0P5JVK9 A0A0P5ANY3 A0A0P6GHE1 A0A0P5D3V4 A0A0P5BZ17 A0A0N8DGC1 A0A0P5ZU81 A0A0P5APW6 A0A0P5M4R8 A0A0P5EYK4 A0A0P5AMF1 A0A0P5LJ49 A0A0P6GRS9 A0A0P6ELQ2 A0A0P5PPC9 A0A0P6FCQ0 A0A0P6ERT9 A0A0P5PME6 A0A0P4ZR35 A0A0P6ES79 A0A0L0C098 A0A0P5JXC3 A0A0P5JJG6 B4MXP4 A0A0P5GU39 A0A0P5GL41 A0A0P5BYV7 A0A0P5BYY4 A0A0P5CB83 A0A0P5BZ82 A0A0P5H0X2 A0A0P5BZ69 A0A0P5H395 A0A0P6EIJ4 A0A164SRT8 A0A0P5K0D6 A0A0P5LDG2 A0A0P5K0E7 A0A1A9YL29 A0A0P5S9U0 A0A0P5SF70 A0A0P5RSS5 A0A0P5RV76 A0A0P5T4L3 A0A0N8CBA5 B4LFN6 A0A1W4W5S9 A0A1W4W6M5 A0A1A9ZFY8 A0A1A9V2Z7 A0A1I8NDE0 A0A0R1DZ34 A0A0R1DZB4 A0A1B0ARA2 A0A1B0FKR1 A0A147BL14
A0A182LW70 A0A1Y1MVJ3 A0A1I8JVT2 A0A2M4CS75 A0A336KKB1 A0A2L1IQ90 K7J800 E0VDI8 X1X3C6 A0A2H8TXV2 A0A2K8JM25 A0A224X4K1 A0A2M4AFQ1 A0A194PCL9 A0A1J1HSS9 A0A0P5TSL7 A0A0P5BYN3 A0A0P5Y7P4 A0A0P6FPD2 A0A0P5PDR7 A0A0P6ALW4 A0A0N8A6F3 A0A0P5C0D4 A0A0N8BRM6 A0A0P5RZV7 A0A0P6DWR4 A0A0P5NJI0 A0A0P5CZJ4 A0A0P6FWK1 A0A0P6DMB0 A0A0N8BMQ8 A0A0P5CUN3 A0A0P5S461 A0A0P6HTE5 A0A0P5WR69 A0A0P5JVK9 A0A0P5ANY3 A0A0P6GHE1 A0A0P5D3V4 A0A0P5BZ17 A0A0N8DGC1 A0A0P5ZU81 A0A0P5APW6 A0A0P5M4R8 A0A0P5EYK4 A0A0P5AMF1 A0A0P5LJ49 A0A0P6GRS9 A0A0P6ELQ2 A0A0P5PPC9 A0A0P6FCQ0 A0A0P6ERT9 A0A0P5PME6 A0A0P4ZR35 A0A0P6ES79 A0A0L0C098 A0A0P5JXC3 A0A0P5JJG6 B4MXP4 A0A0P5GU39 A0A0P5GL41 A0A0P5BYV7 A0A0P5BYY4 A0A0P5CB83 A0A0P5BZ82 A0A0P5H0X2 A0A0P5BZ69 A0A0P5H395 A0A0P6EIJ4 A0A164SRT8 A0A0P5K0D6 A0A0P5LDG2 A0A0P5K0E7 A0A1A9YL29 A0A0P5S9U0 A0A0P5SF70 A0A0P5RSS5 A0A0P5RV76 A0A0P5T4L3 A0A0N8CBA5 B4LFN6 A0A1W4W5S9 A0A1W4W6M5 A0A1A9ZFY8 A0A1A9V2Z7 A0A1I8NDE0 A0A0R1DZ34 A0A0R1DZB4 A0A1B0ARA2 A0A1B0FKR1 A0A147BL14
Pubmed
EMBL
BABH01036590
BABH01036591
BABH01036592
BABH01036593
BABH01036594
BABH01036595
+ More
BABH01036596 BABH01036597 AGBW02013106 OWR43974.1 KQ971312 KYB29318.1 KYB29317.1 GDUN01000114 JAN95805.1 GEZM01019390 JAV89672.1 AXCM01000622 GEZM01019388 JAV89674.1 GGFL01004016 MBW68194.1 UFQS01000216 UFQT01000216 SSX01460.1 SSX21840.1 MF741662 AVD96945.1 DS235078 EEB11444.1 ABLF02039358 ABLF02039360 ABLF02039365 GFXV01006826 MBW18631.1 KY031108 ATU82859.1 GFTR01009000 JAW07426.1 GGFK01006221 MBW39542.1 KQ459606 KPI91036.1 CVRI01000020 CRK90602.1 GDIP01122565 JAL81149.1 GDIP01178028 JAJ45374.1 GDIP01061974 JAM41741.1 GDIQ01046191 JAN48546.1 GDIQ01151650 JAL00076.1 GDIP01027494 JAM76221.1 GDIP01177913 JAJ45489.1 GDIP01177912 JAJ45490.1 GDIQ01151651 JAL00075.1 GDIQ01094817 JAL56909.1 GDIQ01072129 JAN22608.1 GDIQ01151652 JAL00074.1 GDIP01164660 JAJ58742.1 GDIQ01041792 JAN52945.1 GDIQ01090170 JAN04567.1 GDIQ01162595 JAK89130.1 GDIP01165458 JAJ57944.1 GDIQ01092618 JAL59108.1 GDIQ01015694 JAN79043.1 GDIP01082721 JAM20994.1 GDIQ01192933 JAK58792.1 GDIP01196159 JAJ27243.1 GDIQ01050738 GDIQ01033143 JAN61594.1 GDIP01161360 JAJ62042.1 GDIP01177909 JAJ45493.1 GDIP01036479 JAM67236.1 GDIP01039429 JAM64286.1 GDIP01196158 JAJ27244.1 GDIQ01169392 JAK82333.1 GDIQ01268184 JAJ83540.1 GDIP01210858 JAJ12544.1 GDIQ01170241 JAK81484.1 GDIQ01060783 GDIQ01029246 JAN65491.1 GDIQ01094816 GDIQ01060782 JAN33955.1 GDIQ01125358 JAL26368.1 GDIQ01050739 JAN43998.1 GDIQ01072131 JAN22606.1 GDIQ01128408 GDIQ01060784 JAL23318.1 GDIP01210859 JAJ12543.1 GDIQ01072130 JAN22607.1 JRES01001076 KNC25725.1 GDIQ01198031 JAK53694.1 GDIQ01198030 JAK53695.1 CH963876 EDW76813.2 GDIQ01239721 JAK12004.1 GDIQ01239720 JAK12005.1 GDIP01178029 JAJ45373.1 GDIP01178027 JAJ45375.1 GDIP01177911 JAJ45491.1 GDIP01177908 JAJ45494.1 GDIQ01239722 JAK12003.1 GDIP01177910 JAJ45492.1 GDIQ01239719 JAK12006.1 GDIQ01074974 JAN19763.1 LRGB01001937 KZS09882.1 GDIQ01196532 JAK55193.1 GDIQ01196533 JAK55192.1 GDIQ01196534 JAK55191.1 GDIQ01096617 JAL55109.1 GDIQ01096621 JAL55105.1 GDIQ01096616 JAL55110.1 GDIQ01096618 JAL55108.1 GDIQ01096620 JAL55106.1 GDIQ01096619 JAL55107.1 CH940647 EDW70354.2 CM000159 KRK02312.1 KRK02311.1 KRK02313.1 KRK02314.1 JXJN01002295 JXJN01002296 CCAG010023456 CCAG010023457 CCAG010023458 GEGO01004242 JAR91162.1
BABH01036596 BABH01036597 AGBW02013106 OWR43974.1 KQ971312 KYB29318.1 KYB29317.1 GDUN01000114 JAN95805.1 GEZM01019390 JAV89672.1 AXCM01000622 GEZM01019388 JAV89674.1 GGFL01004016 MBW68194.1 UFQS01000216 UFQT01000216 SSX01460.1 SSX21840.1 MF741662 AVD96945.1 DS235078 EEB11444.1 ABLF02039358 ABLF02039360 ABLF02039365 GFXV01006826 MBW18631.1 KY031108 ATU82859.1 GFTR01009000 JAW07426.1 GGFK01006221 MBW39542.1 KQ459606 KPI91036.1 CVRI01000020 CRK90602.1 GDIP01122565 JAL81149.1 GDIP01178028 JAJ45374.1 GDIP01061974 JAM41741.1 GDIQ01046191 JAN48546.1 GDIQ01151650 JAL00076.1 GDIP01027494 JAM76221.1 GDIP01177913 JAJ45489.1 GDIP01177912 JAJ45490.1 GDIQ01151651 JAL00075.1 GDIQ01094817 JAL56909.1 GDIQ01072129 JAN22608.1 GDIQ01151652 JAL00074.1 GDIP01164660 JAJ58742.1 GDIQ01041792 JAN52945.1 GDIQ01090170 JAN04567.1 GDIQ01162595 JAK89130.1 GDIP01165458 JAJ57944.1 GDIQ01092618 JAL59108.1 GDIQ01015694 JAN79043.1 GDIP01082721 JAM20994.1 GDIQ01192933 JAK58792.1 GDIP01196159 JAJ27243.1 GDIQ01050738 GDIQ01033143 JAN61594.1 GDIP01161360 JAJ62042.1 GDIP01177909 JAJ45493.1 GDIP01036479 JAM67236.1 GDIP01039429 JAM64286.1 GDIP01196158 JAJ27244.1 GDIQ01169392 JAK82333.1 GDIQ01268184 JAJ83540.1 GDIP01210858 JAJ12544.1 GDIQ01170241 JAK81484.1 GDIQ01060783 GDIQ01029246 JAN65491.1 GDIQ01094816 GDIQ01060782 JAN33955.1 GDIQ01125358 JAL26368.1 GDIQ01050739 JAN43998.1 GDIQ01072131 JAN22606.1 GDIQ01128408 GDIQ01060784 JAL23318.1 GDIP01210859 JAJ12543.1 GDIQ01072130 JAN22607.1 JRES01001076 KNC25725.1 GDIQ01198031 JAK53694.1 GDIQ01198030 JAK53695.1 CH963876 EDW76813.2 GDIQ01239721 JAK12004.1 GDIQ01239720 JAK12005.1 GDIP01178029 JAJ45373.1 GDIP01178027 JAJ45375.1 GDIP01177911 JAJ45491.1 GDIP01177908 JAJ45494.1 GDIQ01239722 JAK12003.1 GDIP01177910 JAJ45492.1 GDIQ01239719 JAK12006.1 GDIQ01074974 JAN19763.1 LRGB01001937 KZS09882.1 GDIQ01196532 JAK55193.1 GDIQ01196533 JAK55192.1 GDIQ01196534 JAK55191.1 GDIQ01096617 JAL55109.1 GDIQ01096621 JAL55105.1 GDIQ01096616 JAL55110.1 GDIQ01096618 JAL55108.1 GDIQ01096620 JAL55106.1 GDIQ01096619 JAL55107.1 CH940647 EDW70354.2 CM000159 KRK02312.1 KRK02311.1 KRK02313.1 KRK02314.1 JXJN01002295 JXJN01002296 CCAG010023456 CCAG010023457 CCAG010023458 GEGO01004242 JAR91162.1
Proteomes
PRIDE
Pfam
Interpro
IPR002126
Cadherin-like_dom
+ More
IPR001791 Laminin_G
IPR015919 Cadherin-like_sf
IPR020894 Cadherin_CS
IPR013320 ConA-like_dom_sf
IPR001881 EGF-like_Ca-bd_dom
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR018097 EGF_Ca-bd_CS
IPR013032 EGF-like_CS
IPR000742 EGF-like_dom
IPR039808 Cadherin
IPR000233 Cadherin_cytoplasmic-dom
IPR027397 Catenin_binding_dom_sf
IPR023828 Peptidase_S8_Ser-AS
IPR022398 Peptidase_S8_His-AS
IPR036852 Peptidase_S8/S53_dom_sf
IPR000209 Peptidase_S8/S53_dom
IPR015500 Peptidase_S8_subtilisin-rel
IPR001791 Laminin_G
IPR015919 Cadherin-like_sf
IPR020894 Cadherin_CS
IPR013320 ConA-like_dom_sf
IPR001881 EGF-like_Ca-bd_dom
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR018097 EGF_Ca-bd_CS
IPR013032 EGF-like_CS
IPR000742 EGF-like_dom
IPR039808 Cadherin
IPR000233 Cadherin_cytoplasmic-dom
IPR027397 Catenin_binding_dom_sf
IPR023828 Peptidase_S8_Ser-AS
IPR022398 Peptidase_S8_His-AS
IPR036852 Peptidase_S8/S53_dom_sf
IPR000209 Peptidase_S8/S53_dom
IPR015500 Peptidase_S8_subtilisin-rel
Gene 3D
ProteinModelPortal
H9JW17
A0A212ER96
A0A139WMQ2
A0A139WN77
A0A0N8ES92
A0A1Y1MVG4
+ More
A0A182LW70 A0A1Y1MVJ3 A0A1I8JVT2 A0A2M4CS75 A0A336KKB1 A0A2L1IQ90 K7J800 E0VDI8 X1X3C6 A0A2H8TXV2 A0A2K8JM25 A0A224X4K1 A0A2M4AFQ1 A0A194PCL9 A0A1J1HSS9 A0A0P5TSL7 A0A0P5BYN3 A0A0P5Y7P4 A0A0P6FPD2 A0A0P5PDR7 A0A0P6ALW4 A0A0N8A6F3 A0A0P5C0D4 A0A0N8BRM6 A0A0P5RZV7 A0A0P6DWR4 A0A0P5NJI0 A0A0P5CZJ4 A0A0P6FWK1 A0A0P6DMB0 A0A0N8BMQ8 A0A0P5CUN3 A0A0P5S461 A0A0P6HTE5 A0A0P5WR69 A0A0P5JVK9 A0A0P5ANY3 A0A0P6GHE1 A0A0P5D3V4 A0A0P5BZ17 A0A0N8DGC1 A0A0P5ZU81 A0A0P5APW6 A0A0P5M4R8 A0A0P5EYK4 A0A0P5AMF1 A0A0P5LJ49 A0A0P6GRS9 A0A0P6ELQ2 A0A0P5PPC9 A0A0P6FCQ0 A0A0P6ERT9 A0A0P5PME6 A0A0P4ZR35 A0A0P6ES79 A0A0L0C098 A0A0P5JXC3 A0A0P5JJG6 B4MXP4 A0A0P5GU39 A0A0P5GL41 A0A0P5BYV7 A0A0P5BYY4 A0A0P5CB83 A0A0P5BZ82 A0A0P5H0X2 A0A0P5BZ69 A0A0P5H395 A0A0P6EIJ4 A0A164SRT8 A0A0P5K0D6 A0A0P5LDG2 A0A0P5K0E7 A0A1A9YL29 A0A0P5S9U0 A0A0P5SF70 A0A0P5RSS5 A0A0P5RV76 A0A0P5T4L3 A0A0N8CBA5 B4LFN6 A0A1W4W5S9 A0A1W4W6M5 A0A1A9ZFY8 A0A1A9V2Z7 A0A1I8NDE0 A0A0R1DZ34 A0A0R1DZB4 A0A1B0ARA2 A0A1B0FKR1 A0A147BL14
A0A182LW70 A0A1Y1MVJ3 A0A1I8JVT2 A0A2M4CS75 A0A336KKB1 A0A2L1IQ90 K7J800 E0VDI8 X1X3C6 A0A2H8TXV2 A0A2K8JM25 A0A224X4K1 A0A2M4AFQ1 A0A194PCL9 A0A1J1HSS9 A0A0P5TSL7 A0A0P5BYN3 A0A0P5Y7P4 A0A0P6FPD2 A0A0P5PDR7 A0A0P6ALW4 A0A0N8A6F3 A0A0P5C0D4 A0A0N8BRM6 A0A0P5RZV7 A0A0P6DWR4 A0A0P5NJI0 A0A0P5CZJ4 A0A0P6FWK1 A0A0P6DMB0 A0A0N8BMQ8 A0A0P5CUN3 A0A0P5S461 A0A0P6HTE5 A0A0P5WR69 A0A0P5JVK9 A0A0P5ANY3 A0A0P6GHE1 A0A0P5D3V4 A0A0P5BZ17 A0A0N8DGC1 A0A0P5ZU81 A0A0P5APW6 A0A0P5M4R8 A0A0P5EYK4 A0A0P5AMF1 A0A0P5LJ49 A0A0P6GRS9 A0A0P6ELQ2 A0A0P5PPC9 A0A0P6FCQ0 A0A0P6ERT9 A0A0P5PME6 A0A0P4ZR35 A0A0P6ES79 A0A0L0C098 A0A0P5JXC3 A0A0P5JJG6 B4MXP4 A0A0P5GU39 A0A0P5GL41 A0A0P5BYV7 A0A0P5BYY4 A0A0P5CB83 A0A0P5BZ82 A0A0P5H0X2 A0A0P5BZ69 A0A0P5H395 A0A0P6EIJ4 A0A164SRT8 A0A0P5K0D6 A0A0P5LDG2 A0A0P5K0E7 A0A1A9YL29 A0A0P5S9U0 A0A0P5SF70 A0A0P5RSS5 A0A0P5RV76 A0A0P5T4L3 A0A0N8CBA5 B4LFN6 A0A1W4W5S9 A0A1W4W6M5 A0A1A9ZFY8 A0A1A9V2Z7 A0A1I8NDE0 A0A0R1DZ34 A0A0R1DZB4 A0A1B0ARA2 A0A1B0FKR1 A0A147BL14
PDB
6E6B
E-value=4.75382e-51,
Score=515
Ontologies
GO
GO:0005509
GO:0005886
GO:0007156
GO:0016021
GO:0007043
GO:0000902
GO:0009986
GO:0005913
GO:0008092
GO:0034332
GO:0042803
GO:0045296
GO:0098609
GO:0044331
GO:0016339
GO:0016342
GO:0004022
GO:0031254
GO:0007431
GO:0007442
GO:0007424
GO:0042247
GO:0005925
GO:0060269
GO:1902463
GO:0007295
GO:0051491
GO:0007440
GO:0030950
GO:0044877
GO:0050839
GO:0009925
GO:0098858
GO:0004252
GO:0016020
GO:0007155
GO:0005515
GO:0003707
GO:0051537
PANTHER
Topology
Subcellular location
Cell membrane
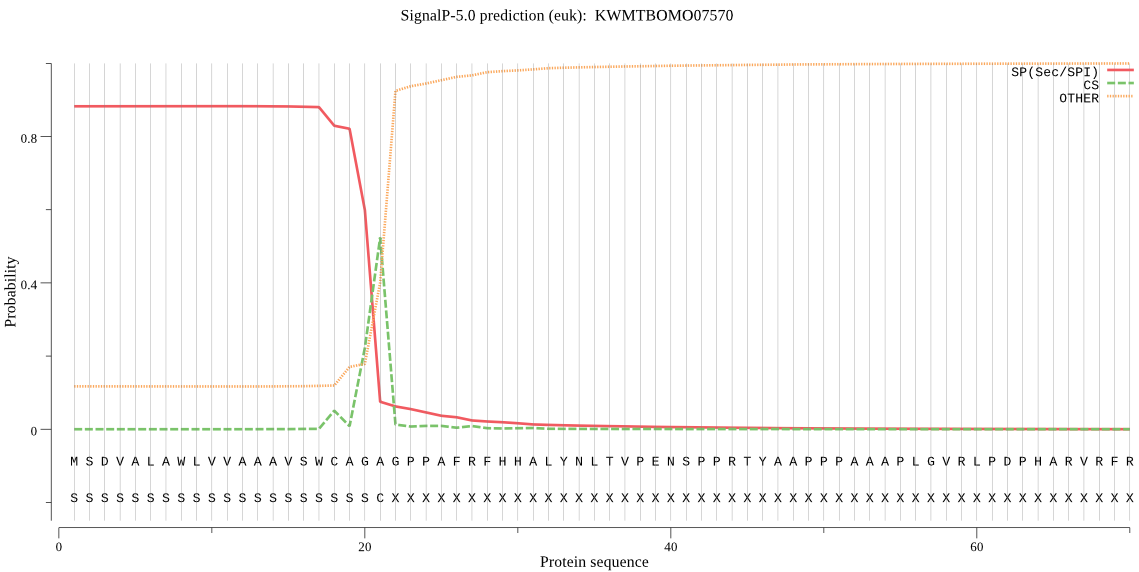
SignalP
Position: 1 - 21,
Likelihood: 0.882123
Length:
2081
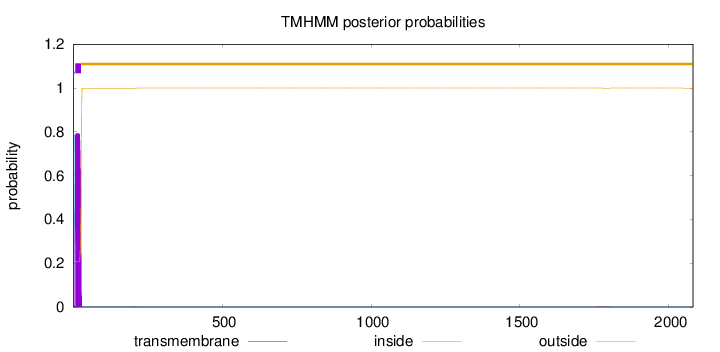
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
16.6403099999999
Exp number, first 60 AAs:
16.60377
Total prob of N-in:
0.79215
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 26
outside
27 - 2081
Population Genetic Test Statistics
Pi
271.868751
Theta
203.853071
Tajima's D
0.961019
CLR
0.128628
CSRT
0.651767411629419
Interpretation
Uncertain
