Gene
KWMTBOMO07563
Pre Gene Modal
BGIBMGA005855
Annotation
PREDICTED:_serine/threonine-protein_kinase_grp_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.23 Mitochondrial Reliability : 1.6
Sequence
CDS
ATGACCGAGTTCGTCGAAGGCTGGGTTGTCGCGCAGGTGCTTGGTGAAGGCGCCTATGGAGAGGTTCGGCTGCTGGTGCACGGCGCGTCGGGCGCGGCGGTGGCGGTGAAGGCGGCCCAGGGCTCGGGCGGCGCGCGGGAGGCAGCCCTGCAGCGCGCGCTGCGGCACCCCCACGTGCTGCGCTGCTTGGGCGAGCGCACGCATCTGCAACATCACTACGTGTTCCTAGAGTACGCGCAGGGCGGCGAGCTCTTCGACAGAATCGAGCCCGACGTGGGCATGGACAGCGACACTGCCCGGCGCTACTGGCGCGAGACGCTGAGCGGCCTGGAGTACCTGCACGCGCGCGGCGTGGCGCACCGCGACATCAAGCCCGAGAACCTGCTGCTCGACCACCACGACCGCGTCAAGATCTCCGACTTTGGCCTGGCCACCGTGTTCCGCCACGGCGGCCGCGAGCGCATGCTGTCCCGCGTGTGCGGCACCCCGCCCTACGCCGCGCCCGAGGTGCTGTCGGCCGCCCAGCGCCCCTACCGCGCCGCGCCCGCCGACCTGTGGTCCGCCGCCGTCGTGCTCGTCGCCATGCTCGCCGGCGAATTGCCGTGGGAGCGTGCGTCGCATTCGGACGCGCGGTACGCGGCGTGGGTGGGGGGCGCGGCGGACGGGGCGGACTCGGCGAGAGCGGGCGCGGCGGACTCTGCGAAGGCGGGCGCGCCGTGGCACAAGCTGCCGCTGCGGTGCGCGTCGCTACTGCGTGCGGCACTGCACCCCGACCCCGCGCGGCGCCCCGCTCTGGCCGACGTGCTCGCGCACCCCTGGACCAGGGACCGCACCTCAGGACCTTTGAGGGACGAACCTCGCGCGTGGTGCTCGCAGCCGTCCTGCGGCCCGGGGCCCGCCTCGCCCAACGAGCTGTCGCAGGACGACATGGAGGCGCTGCTGTGGCACTCGCAGCCCGCGCACGCCGACGAGCTCGTGGTGGAGGCGGCCGTGTCGCAGCGGCTGGTGCGGCGCCGGACGCGCGTGTGGCTGCGCTGCGGCGCGCGCGAGGCGCTGGCGGCGCTGGAGGAGGCGGCGCGGGCGGCGGGGCTGGCGGTGCGGGCGCTGCACGTGCGCGTGCGCGTGCTGGCGGCCGCGGAGCTGCGCGTGCGCGCGTGGGCCGCGCCGCTGCCGCCGGGCCACGAGGCGCGCGCCGTGCTGGAGTTCCGCCGCTCGCGAGGATGCGGGCTGCAGTTCAAGCGCTGCTTCCTGCGGCTCCGCGCGGCGCTGCAGCCGCTGGCCGCGCCCGCGCCCGCGCTGGGGCCCGCGCTGCCGCTCGCCGAGCCCATGCTGCAGGACGCGGCCCCGGGGCCCGCAGATTGA
Protein
MTEFVEGWVVAQVLGEGAYGEVRLLVHGASGAAVAVKAAQGSGGAREAALQRALRHPHVLRCLGERTHLQHHYVFLEYAQGGELFDRIEPDVGMDSDTARRYWRETLSGLEYLHARGVAHRDIKPENLLLDHHDRVKISDFGLATVFRHGGRERMLSRVCGTPPYAAPEVLSAAQRPYRAAPADLWSAAVVLVAMLAGELPWERASHSDARYAAWVGGAADGADSARAGAADSAKAGAPWHKLPLRCASLLRAALHPDPARRPALADVLAHPWTRDRTSGPLRDEPRAWCSQPSCGPGPASPNELSQDDMEALLWHSQPAHADELVVEAAVSQRLVRRRTRVWLRCGAREALAALEEAARAAGLAVRALHVRVRVLAAAELRVRAWAAPLPPGHEARAVLEFRRSRGCGLQFKRCFLRLRAALQPLAAPAPALGPALPLAEPMLQDAAPGPAD
Summary
Similarity
Belongs to the protein kinase superfamily.
Uniprot
H9J8L4
A0A1B6GV01
A0A182QVQ9
A0A182TS63
A0A1B6ILR0
A0A1B6L263
+ More
J9K9N8 A0A0N7ZDQ5 A0A0P6GAN5 A0A0P5RJL9 A0A2S2QEE9 A0A0P4ZII7 A0A0P4WUW8 A0A0P6FH62 A0A0P5DRD4 A0A0P6CN33 E0VNW0 A0A0N8AAG2 E2C708 A0A1B2JLY2 A0A0P5T849 A0A2H8TFR7 R9WA93 A0A0H3V5S2 A0A1E1XSR8 E9FYX6 A0A1E1XFI6 W8BFV7 A1E253 A0A2Z5TZB0 A0A067RWE0 A0A1X7UJ86 A0A2H8TMA4 A0A151IM18 J9K6R9 A0A0M8ZS76 A0A1B0ABD0
J9K9N8 A0A0N7ZDQ5 A0A0P6GAN5 A0A0P5RJL9 A0A2S2QEE9 A0A0P4ZII7 A0A0P4WUW8 A0A0P6FH62 A0A0P5DRD4 A0A0P6CN33 E0VNW0 A0A0N8AAG2 E2C708 A0A1B2JLY2 A0A0P5T849 A0A2H8TFR7 R9WA93 A0A0H3V5S2 A0A1E1XSR8 E9FYX6 A0A1E1XFI6 W8BFV7 A1E253 A0A2Z5TZB0 A0A067RWE0 A0A1X7UJ86 A0A2H8TMA4 A0A151IM18 J9K6R9 A0A0M8ZS76 A0A1B0ABD0
EMBL
BABH01038614
BABH01038615
BABH01038616
GECZ01003495
JAS66274.1
AXCN02000762
+ More
AXCN02000763 GECU01029547 GECU01028909 GECU01019845 JAS78159.1 JAS78797.1 JAS87861.1 GEBQ01022178 JAT17799.1 ABLF02033198 GDRN01010915 JAI67904.1 GDIQ01036221 JAN58516.1 GDIQ01100800 JAL50926.1 GGMS01006369 MBY75572.1 GDIP01212333 JAJ11069.1 GDIP01250494 JAI72907.1 GDIQ01057361 JAN37376.1 GDIP01169367 JAJ54035.1 GDIQ01088929 JAN05808.1 AAZO01004036 DS235354 EEB15066.1 GDIP01166641 JAJ56761.1 GL453255 EFN76280.1 KU958380 ANZ80589.1 GDIP01131945 JAL71769.1 GFXV01000757 MBW12562.1 KC713933 AGN95867.1 KJ735446 AJP08956.1 GFAA01001104 JAU02331.1 GL732527 EFX87696.1 GFAC01001168 JAT98020.1 GAMC01008945 GAMC01008943 GAMC01008942 GAMC01008940 JAB97612.1 EF094480 ABK80717.1 FX985755 BBA93642.1 KK852424 KDR24204.1 GFXV01003492 MBW15297.1 KQ977085 KYN05919.1 ABLF02039873 KQ435879 KOX69887.1
AXCN02000763 GECU01029547 GECU01028909 GECU01019845 JAS78159.1 JAS78797.1 JAS87861.1 GEBQ01022178 JAT17799.1 ABLF02033198 GDRN01010915 JAI67904.1 GDIQ01036221 JAN58516.1 GDIQ01100800 JAL50926.1 GGMS01006369 MBY75572.1 GDIP01212333 JAJ11069.1 GDIP01250494 JAI72907.1 GDIQ01057361 JAN37376.1 GDIP01169367 JAJ54035.1 GDIQ01088929 JAN05808.1 AAZO01004036 DS235354 EEB15066.1 GDIP01166641 JAJ56761.1 GL453255 EFN76280.1 KU958380 ANZ80589.1 GDIP01131945 JAL71769.1 GFXV01000757 MBW12562.1 KC713933 AGN95867.1 KJ735446 AJP08956.1 GFAA01001104 JAU02331.1 GL732527 EFX87696.1 GFAC01001168 JAT98020.1 GAMC01008945 GAMC01008943 GAMC01008942 GAMC01008940 JAB97612.1 EF094480 ABK80717.1 FX985755 BBA93642.1 KK852424 KDR24204.1 GFXV01003492 MBW15297.1 KQ977085 KYN05919.1 ABLF02039873 KQ435879 KOX69887.1
Proteomes
PRIDE
Pfam
PF00069 Pkinase
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
H9J8L4
A0A1B6GV01
A0A182QVQ9
A0A182TS63
A0A1B6ILR0
A0A1B6L263
+ More
J9K9N8 A0A0N7ZDQ5 A0A0P6GAN5 A0A0P5RJL9 A0A2S2QEE9 A0A0P4ZII7 A0A0P4WUW8 A0A0P6FH62 A0A0P5DRD4 A0A0P6CN33 E0VNW0 A0A0N8AAG2 E2C708 A0A1B2JLY2 A0A0P5T849 A0A2H8TFR7 R9WA93 A0A0H3V5S2 A0A1E1XSR8 E9FYX6 A0A1E1XFI6 W8BFV7 A1E253 A0A2Z5TZB0 A0A067RWE0 A0A1X7UJ86 A0A2H8TMA4 A0A151IM18 J9K6R9 A0A0M8ZS76 A0A1B0ABD0
J9K9N8 A0A0N7ZDQ5 A0A0P6GAN5 A0A0P5RJL9 A0A2S2QEE9 A0A0P4ZII7 A0A0P4WUW8 A0A0P6FH62 A0A0P5DRD4 A0A0P6CN33 E0VNW0 A0A0N8AAG2 E2C708 A0A1B2JLY2 A0A0P5T849 A0A2H8TFR7 R9WA93 A0A0H3V5S2 A0A1E1XSR8 E9FYX6 A0A1E1XFI6 W8BFV7 A1E253 A0A2Z5TZB0 A0A067RWE0 A0A1X7UJ86 A0A2H8TMA4 A0A151IM18 J9K6R9 A0A0M8ZS76 A0A1B0ABD0
PDB
3JVS
E-value=1.45681e-55,
Score=548
Ontologies
GO
Topology
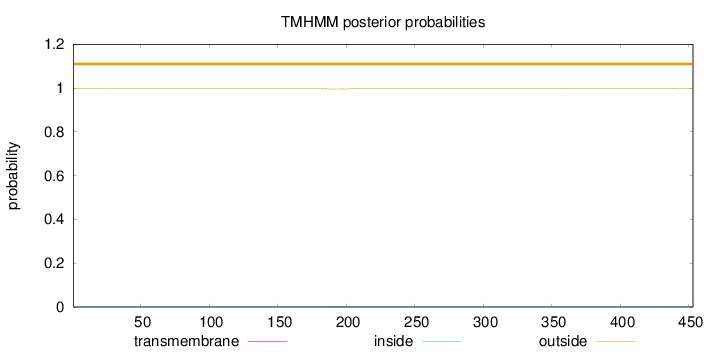
Length:
453
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06522
Exp number, first 60 AAs:
0.01171
Total prob of N-in:
0.00359
outside
1 - 453
Population Genetic Test Statistics
Pi
6.906417
Theta
7.115158
Tajima's D
0.157242
CLR
0.664063
CSRT
0.432878356082196
Interpretation
Uncertain
