Gene
KWMTBOMO07562
Annotation
PREDICTED:_protein_phosphatase_1_regulatory_subunit_14B_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.416
Sequence
CDS
ATGTCGCTGTTCTTCAAGTTCAGAAGCAACCGAGGCAGCGGCGCCGGTGCGGCTGCGTGCGGCATGGAGTGCGGAGTGGCGGCGAGCGGCTTCCCCGCGCCCGCACCCCGCGCACACCCCGCCCTCTCCCCGACAGAGCGCTCCCCCGCCAAGGCCGGACTGCACGTCAACTTCAGCGATAAGGGAGAGGTAAAAGAGCGACGCGAGAAGTTCCTAACGGCGAAGTATGGCAGCCATCAGATGGCACTGATACGCAAGCGACTCGCGGTCGAGATGTGGCTCTACGACGAGCTGCAGAAGCTCTACGAGATACCGAAGGAGAGCAGCGGGGCGGCGGGCGCGCAGGAGGTGGAGGTGGACATCGACGAGCTCCTGGACATGGACACCGACGACTCGCGGCGGCGCCACCTCGCGACATTGCTAGCGGATGCCAAAAAGCCACAAAAGGACGTCCACAAGTTTATCAACGAATTATTAGACAAGGCTAAGACTCTGTAA
Protein
MSLFFKFRSNRGSGAGAAACGMECGVAASGFPAPAPRAHPALSPTERSPAKAGLHVNFSDKGEVKERREKFLTAKYGSHQMALIRKRLAVEMWLYDELQKLYEIPKESSGAAGAQEVEVDIDELLDMDTDDSRRRHLATLLADAKKPQKDVHKFINELLDKAKTL
Summary
Uniprot
A0A212F6B0
A0A0N1PG86
S4NYB9
A0A2W1BUL6
H9J8L0
A0A2H1WL13
+ More
A0A1Y1K176 D6X289 A0A1B6FF05 A0A1B6HPK8 A0A1B6D1L8 A0A1B6IPS3 A0A1L8DKY2 A0A1B6KL90 A0A0N0U564 A0A0C9RCI4 A0A154NXR0 A0A2A3E5M0 A0A088ASZ5 A0A0A9XYZ6 A0A023F903 A0A0L7RAV1 R4G3R2 A0A1Q3FIY7 B0WBN1 U5EQV0 A0A0K8S419 Q175K5 A0A182GJT1 A0A310SFJ1 T1E2M6 A0A182RBR1 A0A182NE77 A0A182V884 A0A182WTD0 Q7PN37 A0A182W9U0 U4TYT5 T1DQU0 W5JBN9 A0A1J1ISZ8 A0A232EGA9 K7ISN1 A0A336MEM5 T1P9U5 A0A1B0CDC6 A0A1I8NMM9 B4KKN4 A0A0M4ES93 A0A1A9XZI8 A0A1A9UMJ4 A0A1B0B903 A0A1A9Z7S8 A0A1A9WZW8 A0A3B0JN35 B4GSZ8 Q29KQ3 B4MU57 A0A034VTY0 A0A0K8W7N3 B3MJV5 B4P0U2 B4HWV3 B3N537 B4Q9T3 Q9VKQ5 W8C4G5 A0A0A1XFI1 A0A1W4W550 B4LS81 B4JE51 A0A1B0G5Z1 A0A226DBY0 A0A2H8TG25 A0A2S2QPK4 A0A182PMC3 A0A182JGP3 A0A0K8VU19
A0A1Y1K176 D6X289 A0A1B6FF05 A0A1B6HPK8 A0A1B6D1L8 A0A1B6IPS3 A0A1L8DKY2 A0A1B6KL90 A0A0N0U564 A0A0C9RCI4 A0A154NXR0 A0A2A3E5M0 A0A088ASZ5 A0A0A9XYZ6 A0A023F903 A0A0L7RAV1 R4G3R2 A0A1Q3FIY7 B0WBN1 U5EQV0 A0A0K8S419 Q175K5 A0A182GJT1 A0A310SFJ1 T1E2M6 A0A182RBR1 A0A182NE77 A0A182V884 A0A182WTD0 Q7PN37 A0A182W9U0 U4TYT5 T1DQU0 W5JBN9 A0A1J1ISZ8 A0A232EGA9 K7ISN1 A0A336MEM5 T1P9U5 A0A1B0CDC6 A0A1I8NMM9 B4KKN4 A0A0M4ES93 A0A1A9XZI8 A0A1A9UMJ4 A0A1B0B903 A0A1A9Z7S8 A0A1A9WZW8 A0A3B0JN35 B4GSZ8 Q29KQ3 B4MU57 A0A034VTY0 A0A0K8W7N3 B3MJV5 B4P0U2 B4HWV3 B3N537 B4Q9T3 Q9VKQ5 W8C4G5 A0A0A1XFI1 A0A1W4W550 B4LS81 B4JE51 A0A1B0G5Z1 A0A226DBY0 A0A2H8TG25 A0A2S2QPK4 A0A182PMC3 A0A182JGP3 A0A0K8VU19
Pubmed
22118469
26354079
23622113
28756777
19121390
28004739
+ More
18362917 19820115 25401762 26823975 25474469 17510324 26483478 24330624 12364791 14747013 17210077 23537049 20920257 23761445 28648823 20075255 25315136 17994087 15632085 23185243 25348373 17550304 22936249 10731132 12537568 12537572 12537573 12537574 12974676 16110336 17569856 17569867 26109357 26109356 24495485 25830018 18057021
18362917 19820115 25401762 26823975 25474469 17510324 26483478 24330624 12364791 14747013 17210077 23537049 20920257 23761445 28648823 20075255 25315136 17994087 15632085 23185243 25348373 17550304 22936249 10731132 12537568 12537572 12537573 12537574 12974676 16110336 17569856 17569867 26109357 26109356 24495485 25830018 18057021
EMBL
AGBW02010072
OWR49254.1
KQ460642
KPJ13354.1
GAIX01013860
JAA78700.1
+ More
KZ149931 PZC77324.1 BABH01038617 BABH01038618 ODYU01009321 SOQ53656.1 GEZM01097379 GEZM01097378 JAV54238.1 KQ971371 EFA10228.2 GECZ01031496 GECZ01020977 JAS38273.1 JAS48792.1 GECU01031088 JAS76618.1 GEDC01017720 GEDC01005947 JAS19578.1 JAS31351.1 GECU01018819 JAS88887.1 GFDF01006968 JAV07116.1 GEBQ01027760 JAT12217.1 KQ435798 KOX73451.1 GBYB01006020 JAG75787.1 KQ434781 KZC04465.1 KZ288361 PBC26995.1 GBHO01017563 GBHO01017562 GBRD01017801 GDHC01020106 GDHC01007044 GDHC01000500 JAG26041.1 JAG26042.1 JAG48026.1 JAP98522.1 JAQ11585.1 JAQ18129.1 GBBI01000775 JAC17937.1 KQ414618 KOC67994.1 GAHY01001441 JAA76069.1 GFDL01007557 JAV27488.1 DS231880 EDS42566.1 GANO01004052 JAB55819.1 GBRD01017800 JAG48027.1 CH477398 EAT41797.1 JXUM01013688 JXUM01013689 JXUM01013690 KQ560382 KXJ82695.1 KQ760498 OAD60088.1 GALA01000569 JAA94283.1 AAAB01008964 EAA12834.2 KB631865 ERL86784.1 GAMD01002264 JAA99326.1 ADMH02001962 GGFL01004437 ETN60184.1 MBW68615.1 CVRI01000058 CRL02858.1 NNAY01004813 OXU17381.1 AAZX01011284 UFQS01001088 UFQT01001088 SSX08931.1 SSX28842.1 KA645531 AFP60160.1 AJWK01007660 AJWK01007661 AJWK01007662 CH933807 EDW12698.1 CP012523 ALC40162.1 JXJN01010178 JXJN01010179 OUUW01000006 SPP81752.1 CH479189 EDW25507.1 CH379061 EAL33121.1 KRT04754.1 KRT04755.1 CH963852 EDW75646.1 GAKP01012196 JAC46756.1 GDHF01031228 GDHF01021923 GDHF01005227 JAI21086.1 JAI30391.1 JAI47087.1 CH902620 EDV32410.1 CM000157 EDW87987.1 CH480818 EDW52498.1 CH954177 EDV58982.1 CM000361 CM002910 EDX04600.1 KMY89616.1 AE014134 AY050669 AY095067 KX531835 AAF53006.1 AAL25827.1 AAM11395.1 AAN10761.1 ANY27645.1 GAMC01002317 GAMC01002316 JAC04239.1 GBXI01010791 GBXI01009595 GBXI01004128 JAD03501.1 JAD04697.1 JAD10164.1 CH940649 EDW64767.1 KRF81849.1 CH916368 EDW03571.1 CCAG010022414 CCAG010022415 CCAG010022416 LNIX01000025 OXA42719.1 GFXV01001195 MBW13000.1 GGMS01010400 MBY79603.1 GDHF01030551 GDHF01009938 JAI21763.1 JAI42376.1
KZ149931 PZC77324.1 BABH01038617 BABH01038618 ODYU01009321 SOQ53656.1 GEZM01097379 GEZM01097378 JAV54238.1 KQ971371 EFA10228.2 GECZ01031496 GECZ01020977 JAS38273.1 JAS48792.1 GECU01031088 JAS76618.1 GEDC01017720 GEDC01005947 JAS19578.1 JAS31351.1 GECU01018819 JAS88887.1 GFDF01006968 JAV07116.1 GEBQ01027760 JAT12217.1 KQ435798 KOX73451.1 GBYB01006020 JAG75787.1 KQ434781 KZC04465.1 KZ288361 PBC26995.1 GBHO01017563 GBHO01017562 GBRD01017801 GDHC01020106 GDHC01007044 GDHC01000500 JAG26041.1 JAG26042.1 JAG48026.1 JAP98522.1 JAQ11585.1 JAQ18129.1 GBBI01000775 JAC17937.1 KQ414618 KOC67994.1 GAHY01001441 JAA76069.1 GFDL01007557 JAV27488.1 DS231880 EDS42566.1 GANO01004052 JAB55819.1 GBRD01017800 JAG48027.1 CH477398 EAT41797.1 JXUM01013688 JXUM01013689 JXUM01013690 KQ560382 KXJ82695.1 KQ760498 OAD60088.1 GALA01000569 JAA94283.1 AAAB01008964 EAA12834.2 KB631865 ERL86784.1 GAMD01002264 JAA99326.1 ADMH02001962 GGFL01004437 ETN60184.1 MBW68615.1 CVRI01000058 CRL02858.1 NNAY01004813 OXU17381.1 AAZX01011284 UFQS01001088 UFQT01001088 SSX08931.1 SSX28842.1 KA645531 AFP60160.1 AJWK01007660 AJWK01007661 AJWK01007662 CH933807 EDW12698.1 CP012523 ALC40162.1 JXJN01010178 JXJN01010179 OUUW01000006 SPP81752.1 CH479189 EDW25507.1 CH379061 EAL33121.1 KRT04754.1 KRT04755.1 CH963852 EDW75646.1 GAKP01012196 JAC46756.1 GDHF01031228 GDHF01021923 GDHF01005227 JAI21086.1 JAI30391.1 JAI47087.1 CH902620 EDV32410.1 CM000157 EDW87987.1 CH480818 EDW52498.1 CH954177 EDV58982.1 CM000361 CM002910 EDX04600.1 KMY89616.1 AE014134 AY050669 AY095067 KX531835 AAF53006.1 AAL25827.1 AAM11395.1 AAN10761.1 ANY27645.1 GAMC01002317 GAMC01002316 JAC04239.1 GBXI01010791 GBXI01009595 GBXI01004128 JAD03501.1 JAD04697.1 JAD10164.1 CH940649 EDW64767.1 KRF81849.1 CH916368 EDW03571.1 CCAG010022414 CCAG010022415 CCAG010022416 LNIX01000025 OXA42719.1 GFXV01001195 MBW13000.1 GGMS01010400 MBY79603.1 GDHF01030551 GDHF01009938 JAI21763.1 JAI42376.1
Proteomes
UP000007151
UP000053240
UP000005204
UP000007266
UP000053105
UP000076502
+ More
UP000242457 UP000005203 UP000053825 UP000002320 UP000008820 UP000069940 UP000249989 UP000075900 UP000075884 UP000075903 UP000076407 UP000007062 UP000075920 UP000030742 UP000000673 UP000183832 UP000215335 UP000002358 UP000095301 UP000092461 UP000095300 UP000009192 UP000092553 UP000092443 UP000078200 UP000092460 UP000092445 UP000091820 UP000268350 UP000008744 UP000001819 UP000007798 UP000007801 UP000002282 UP000001292 UP000008711 UP000000304 UP000000803 UP000192221 UP000008792 UP000001070 UP000092444 UP000198287 UP000075885 UP000075880
UP000242457 UP000005203 UP000053825 UP000002320 UP000008820 UP000069940 UP000249989 UP000075900 UP000075884 UP000075903 UP000076407 UP000007062 UP000075920 UP000030742 UP000000673 UP000183832 UP000215335 UP000002358 UP000095301 UP000092461 UP000095300 UP000009192 UP000092553 UP000092443 UP000078200 UP000092460 UP000092445 UP000091820 UP000268350 UP000008744 UP000001819 UP000007798 UP000007801 UP000002282 UP000001292 UP000008711 UP000000304 UP000000803 UP000192221 UP000008792 UP000001070 UP000092444 UP000198287 UP000075885 UP000075880
Pfam
PF05361 PP1_inhibitor
SUPFAM
SSF81790
SSF81790
Gene 3D
ProteinModelPortal
A0A212F6B0
A0A0N1PG86
S4NYB9
A0A2W1BUL6
H9J8L0
A0A2H1WL13
+ More
A0A1Y1K176 D6X289 A0A1B6FF05 A0A1B6HPK8 A0A1B6D1L8 A0A1B6IPS3 A0A1L8DKY2 A0A1B6KL90 A0A0N0U564 A0A0C9RCI4 A0A154NXR0 A0A2A3E5M0 A0A088ASZ5 A0A0A9XYZ6 A0A023F903 A0A0L7RAV1 R4G3R2 A0A1Q3FIY7 B0WBN1 U5EQV0 A0A0K8S419 Q175K5 A0A182GJT1 A0A310SFJ1 T1E2M6 A0A182RBR1 A0A182NE77 A0A182V884 A0A182WTD0 Q7PN37 A0A182W9U0 U4TYT5 T1DQU0 W5JBN9 A0A1J1ISZ8 A0A232EGA9 K7ISN1 A0A336MEM5 T1P9U5 A0A1B0CDC6 A0A1I8NMM9 B4KKN4 A0A0M4ES93 A0A1A9XZI8 A0A1A9UMJ4 A0A1B0B903 A0A1A9Z7S8 A0A1A9WZW8 A0A3B0JN35 B4GSZ8 Q29KQ3 B4MU57 A0A034VTY0 A0A0K8W7N3 B3MJV5 B4P0U2 B4HWV3 B3N537 B4Q9T3 Q9VKQ5 W8C4G5 A0A0A1XFI1 A0A1W4W550 B4LS81 B4JE51 A0A1B0G5Z1 A0A226DBY0 A0A2H8TG25 A0A2S2QPK4 A0A182PMC3 A0A182JGP3 A0A0K8VU19
A0A1Y1K176 D6X289 A0A1B6FF05 A0A1B6HPK8 A0A1B6D1L8 A0A1B6IPS3 A0A1L8DKY2 A0A1B6KL90 A0A0N0U564 A0A0C9RCI4 A0A154NXR0 A0A2A3E5M0 A0A088ASZ5 A0A0A9XYZ6 A0A023F903 A0A0L7RAV1 R4G3R2 A0A1Q3FIY7 B0WBN1 U5EQV0 A0A0K8S419 Q175K5 A0A182GJT1 A0A310SFJ1 T1E2M6 A0A182RBR1 A0A182NE77 A0A182V884 A0A182WTD0 Q7PN37 A0A182W9U0 U4TYT5 T1DQU0 W5JBN9 A0A1J1ISZ8 A0A232EGA9 K7ISN1 A0A336MEM5 T1P9U5 A0A1B0CDC6 A0A1I8NMM9 B4KKN4 A0A0M4ES93 A0A1A9XZI8 A0A1A9UMJ4 A0A1B0B903 A0A1A9Z7S8 A0A1A9WZW8 A0A3B0JN35 B4GSZ8 Q29KQ3 B4MU57 A0A034VTY0 A0A0K8W7N3 B3MJV5 B4P0U2 B4HWV3 B3N537 B4Q9T3 Q9VKQ5 W8C4G5 A0A0A1XFI1 A0A1W4W550 B4LS81 B4JE51 A0A1B0G5Z1 A0A226DBY0 A0A2H8TG25 A0A2S2QPK4 A0A182PMC3 A0A182JGP3 A0A0K8VU19
Ontologies
GO
PANTHER
Topology
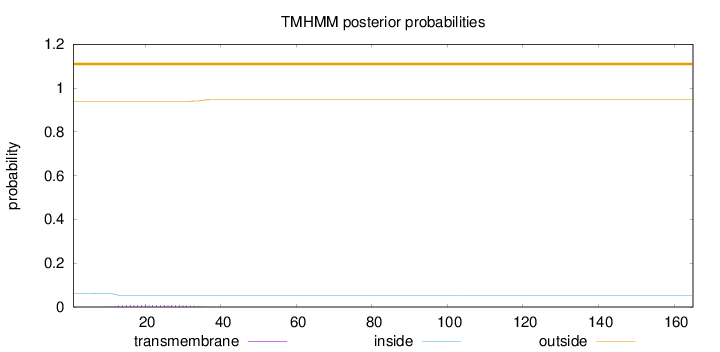
Length:
165
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.17923
Exp number, first 60 AAs:
0.17923
Total prob of N-in:
0.06136
outside
1 - 165
Population Genetic Test Statistics
Pi
3.2769
Theta
5.670198
Tajima's D
-1.355687
CLR
0.48513
CSRT
0.0746462676866157
Interpretation
Uncertain
