Gene
KWMTBOMO07561
Annotation
PREDICTED:_guanine_nucleotide_binding_protein_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.769
Sequence
CDS
ATGGAGAGTCGCGCGTGGGTGTGGTGGCTGGGAGTGGCGGCGCTGGCGAGCGCGCGCGAGCTGGACCCGGCCGCCTGCGTGGCGCGCTTCGACGTGCAGCGAGACAAGATCATCCGCACCGAGGAGTCCCGCGAGATGGGCGCCCGCTACCTCTCCGAGCTTGACGTGGGCACGAGAGCGGAATGCCTGCGACTCTGCTGTGAAACTGAAGCCTGCGACGTCTTCGTCTATGAGGAAAAGGGTCCCGGCAGCTGCTACTTGTTCAAGTGTGGACCGCCCGAAGACTTCCGCTGCAAGTTCACGGCTCACAGTAACTTCAGTAGCGGAGTGCTGGCGCTGAGTCGCCGCCTGGCCGAGCTGCAGGACCAGGAGCAACGCGCCCACCACGAGCGGGAACTCGCCAACCTCAGTCACATGTTGGGAACGCGCTACAGGAGCGGGCCGGGCACCGCCGCGAGCACCAGCACGACGCCCGCGCCGCCGCCCCCGCCCCCGCCCCGCACGAACTTACCTCTCCAGTCTCCGGCCCCCACGCCGGGTCGATGCCGCTGGAACCAGTTCGAGTGTCGTCGCGGCGGGGAGTGCGTCGCGGTGTACAACGCGTGCGACGGCGTGCCGCAGTGCGCCGACGGCAGCGACGAGCCGCCCCAGCTGTCGTGTCCATCCCCCCCAACCGCCCCCTCCGCCCTCCCCATCACCACCGTGCCGACCACCACGCGGCACGCACCACCGATGCAGGACAGCATGGACCTGAGCGTGGGCAGCGACAGTCTCGTGGACGGAGACCGGTGGCCGCACCGGCTGACGCAGGCTCAGCCGCCACACAGATACACGGAGTCCGGTCCGAGCTCTCATATCTTCAGTCACAAGGGCGGGTTGCTTCAGGAGCCGAGCGGCGACGCCCAGTTCCCCGCCTTCGAGCCCCCCTGGCAGCGACGGCCTTGGGGATCGCAAGCCGGTGTGGGAGGGATGGAGGAGCGTGGGTGGGGGGCAGGATACCCGCGGCTCGCGTGGCCCGAGCCCGACCTGCGCCGCGCCTGGCCCGTTCCGCAGCAGCGACAAGACACGGAGATGCAGATGTATATGCCTGCGAAGACAATGCCCGAGGTGCCGATAGTGTACTCTAGTCAGCAACAAACTCTCCGCGACGGACCTAAGAAGGGCCTGGAGCTGCCACCCCCCACCGCCCCGCCACCCTCCCCTGCTCCCGATCTGAGAGAGGAGAACAAGCCGCCCCTACTTGAGAATCATACTAAGAAGAAACCTATTAAAGAAATAATACAGACGGCAGAGTCGAACAAGGCGGTCGCGCGGGAGCCCGGCGGCGCGGCGTCGGCCCGGCGCCACCCCGCGCCCCCGCCCCCGCTCGACGACCACGACGGCCTCAGTGAGCATCCCCCCGCCGCCGTGCTGCTGCTAGTGCTCGGCGTGTCGCTGACGGGCGCGCTGGCGGGCATGCTGGCGTGTCGTGCGCGCGCGGCGCGGCGCCGGCTGCGACGCAAGTCCGCGCTGGCGCACGACGCGGACTACCTCGTCAACGGGATGTACCTGTAG
Protein
MESRAWVWWLGVAALASARELDPAACVARFDVQRDKIIRTEESREMGARYLSELDVGTRAECLRLCCETEACDVFVYEEKGPGSCYLFKCGPPEDFRCKFTAHSNFSSGVLALSRRLAELQDQEQRAHHERELANLSHMLGTRYRSGPGTAASTSTTPAPPPPPPPRTNLPLQSPAPTPGRCRWNQFECRRGGECVAVYNACDGVPQCADGSDEPPQLSCPSPPTAPSALPITTVPTTTRHAPPMQDSMDLSVGSDSLVDGDRWPHRLTQAQPPHRYTESGPSSHIFSHKGGLLQEPSGDAQFPAFEPPWQRRPWGSQAGVGGMEERGWGAGYPRLAWPEPDLRRAWPVPQQRQDTEMQMYMPAKTMPEVPIVYSSQQQTLRDGPKKGLELPPPTAPPPSPAPDLREENKPPLLENHTKKKPIKEIIQTAESNKAVAREPGGAASARRHPAPPPPLDDHDGLSEHPPAAVLLLVLGVSLTGALAGMLACRARAARRRLRRKSALAHDADYLVNGMYL
Summary
Uniprot
Pubmed
Proteomes
PRIDE
Interpro
SUPFAM
SSF57424
SSF57424
Gene 3D
CDD
ProteinModelPortal
PDB
2MSX
E-value=0.000994628,
Score=101
Ontologies
GO
Topology
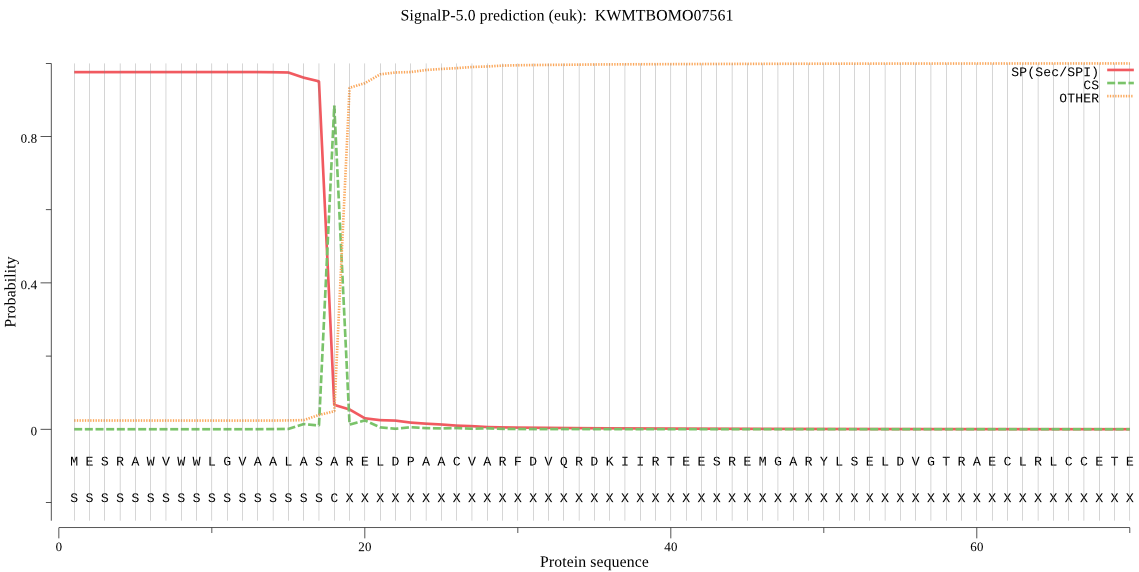
SignalP
Position: 1 - 18,
Likelihood: 0.975640
Length:
517
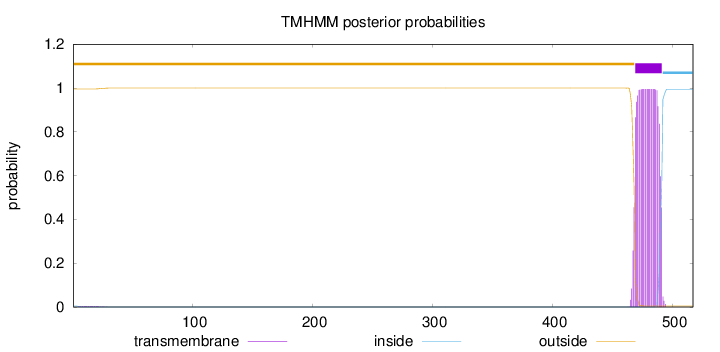
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.47942
Exp number, first 60 AAs:
0.08447
Total prob of N-in:
0.00457
outside
1 - 468
TMhelix
469 - 491
inside
492 - 517
Population Genetic Test Statistics
Pi
2.017732
Theta
5.745484
Tajima's D
-1.730906
CLR
36.675421
CSRT
0.0305984700764962
Interpretation
Uncertain
