Gene
KWMTBOMO07559 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005859
Annotation
PREDICTED:_chitinase-3-like_protein_1_[Papilio_xuthus]
Location in the cell
Extracellular Reliability : 1.361 PlasmaMembrane Reliability : 1.04
Sequence
CDS
ATGCGTTTAGATTACGATGGAATTCTGACGAAAGCGAGCCTTTTCATAGTCCTTGGAACTGCTGTATACATGTCGATGCTAACTCTGGTATATAGTGGTCAGCTGCATTCCAAAAGTCAGGATAATGTGTTCTTCATCCCAGTTACATCAGACATCAAAAGTAATACAGTAGTGTCTTGTTATTACAACACACCTGATAATGATGGCAAACAACTATTGCCAGCCAGTATTGACCCTCACCTGTGCACACACATCAATGTTGCATTTGCAAGAGTCATCGACAAAAAGATACATCTAGATGAATATCAATACCAAACTATAACTGAATTGTGCAAACTCAAACAGGAAAATCCTGCCCTTAAAGTATTGATTTCAGTGGGAGGTGCTGGAGAAGCTTCTGGTTTTAGAGACATGGTTGCCAATCATGCTTCTAGGAAAATTTTTATCAAATCAATCAAAACTATTCTAAGAAATTACAAATTAGATGGAATAGATTTAGACTGGGAGTTTCCAGAAGTGCACAAGCTTTATAAAATGAAAGGAAGTCGCGAGAGGCAACACTTCTCCCAGTTACTGCGTGAAATCAGAATGGAATATTTGAGGGAAAAGCGAGATTACCTGCTGACTGTAGCTGTTGCTGCTCCAGAGGCAATAGTTGATGTTTCCTACGATGTGCAGCAGATTAATAATTATGTAGACTATGTTAACATCATGACATATGACTACCACTATTATACTAAATACACTCCATTCACTGGCTTCAACTCCCCATTATTCTCAAGACCCTCGGAAAGTTTGTACATGGGCACACTTAATGTTAATTACACAGTCAACATGTATTTAAATAAAGGATTGGATCGAGATAAGATAGTTGTTGGAATACCAACATATGGACATTCATTTACTCTAGTGAATGCAGACAACCCAAATATTGGCAGTCCAGCTTCAGGCTACGGATCTCTAGGTTCACTAGGCTTTGTTAATTACCCAGATGTTTGCTTATTTGTAGCCAACAGTAATGTTAGAGTGATATTTGATGATCACGCAAAAGTGCCTTACTTATATAAACAGAAAGAATGGACCTCATTTGACTCAGCTCACAGTGTCATGGAGAAGGCAAAATATATAAAAGACAATAGACTACGAGGTGCTATGGTGTATTCATTAAATGCTGATGACTATGAAGGAAAATGTACTAGTAAAACAAATAACACCATACTGTTCCCTCTTGTTCAATCAATTAGGGACACTCTCATAACCAAAAGCACACAAGATGTTATACTTGTCTAA
Protein
MRLDYDGILTKASLFIVLGTAVYMSMLTLVYSGQLHSKSQDNVFFIPVTSDIKSNTVVSCYYNTPDNDGKQLLPASIDPHLCTHINVAFARVIDKKIHLDEYQYQTITELCKLKQENPALKVLISVGGAGEASGFRDMVANHASRKIFIKSIKTILRNYKLDGIDLDWEFPEVHKLYKMKGSRERQHFSQLLREIRMEYLREKRDYLLTVAVAAPEAIVDVSYDVQQINNYVDYVNIMTYDYHYYTKYTPFTGFNSPLFSRPSESLYMGTLNVNYTVNMYLNKGLDRDKIVVGIPTYGHSFTLVNADNPNIGSPASGYGSLGSLGFVNYPDVCLFVANSNVRVIFDDHAKVPYLYKQKEWTSFDSAHSVMEKAKYIKDNRLRGAMVYSLNADDYEGKCTSKTNNTILFPLVQSIRDTLITKSTQDVILV
Summary
Similarity
Belongs to the glycosyl hydrolase 18 family.
Uniprot
H9J8L7
A0A2H1WCM1
A0A0N1IFU3
A0A194PU30
A0A2W1BQG7
A0A2A4K0C3
+ More
A0A0L7KVP9 A0A1L8E1M5 A0A1Y1JQQ8 A0A0T6AYA6 A0A0A1WME4 A0A0L0BUY6 A0A0K8U0Z8 U4TTQ9 N6SVA0 W8BXL9 A0A034VVU5 A0A2P0P9G2 A0A2J7QUX9 A0A1I8NSV0 A0A182V6E0 A0A026WFT6 Q7QFE2 A0A0K8TLZ9 A0A182W5G7 A0A067QY08 A0A182M3W6 A0A232F6F5 K7IT18 A0A310SRX5 A0A182N1A3 A0A0J7KR08 A0A2A3E0P8 V9IIB8 E2A7N2 A0A088A858 A0A182T4N0 A0A0M9A3Y2 E9I9P7 E2BWT8 A0A1J1HR99 A0A182FJA1 A0A1I8MB77 A0A195EVA5 A0A182PBK0 A0A151J0Q1 A0A182QCP1 A0A158NAE7 A0A151I0U7 A0A195CJY5 A0A1A9XXZ7 A0A0C9RT37 A0A182IUW3 A0A1B0BX17 A0A182XXU8 A0A1A9WA71 A0A084VLE5 A0A1A9Z8J7 A0A1B0FIC8 A0A1A9UM59 B4M6Y8 A0A1Q3FUW7 A0A182TNA2 B3MS63 A0A154P3X4 W5JST5 A0A3B0JDW9 B4PZ92 A0A0M3QZE6 Q16UV6 A0A1S4FND0 B4JXI2 A0A151X588 A0A182XDT0 B3NXX0 Q9W3V1 A0A1W4UVS0 A0A1B6E7G7 A0A1B6DPY5 B4L512 A0A1B6MSA9 A0A0C5CUD5 B4MT40 Q29I77 A0A0P4VGW2 A0A1B6FFK2 T1I6U9 A0A1B6FU96 A0A2S2RBL2 F4WMI9 B4GWJ5 A0A0A9VRR7 A0A0K8TAJ9 X1WJN6 A0A146L7K0 A0A146LM60 A0A023EZV2 A0A3Q0IW64
A0A0L7KVP9 A0A1L8E1M5 A0A1Y1JQQ8 A0A0T6AYA6 A0A0A1WME4 A0A0L0BUY6 A0A0K8U0Z8 U4TTQ9 N6SVA0 W8BXL9 A0A034VVU5 A0A2P0P9G2 A0A2J7QUX9 A0A1I8NSV0 A0A182V6E0 A0A026WFT6 Q7QFE2 A0A0K8TLZ9 A0A182W5G7 A0A067QY08 A0A182M3W6 A0A232F6F5 K7IT18 A0A310SRX5 A0A182N1A3 A0A0J7KR08 A0A2A3E0P8 V9IIB8 E2A7N2 A0A088A858 A0A182T4N0 A0A0M9A3Y2 E9I9P7 E2BWT8 A0A1J1HR99 A0A182FJA1 A0A1I8MB77 A0A195EVA5 A0A182PBK0 A0A151J0Q1 A0A182QCP1 A0A158NAE7 A0A151I0U7 A0A195CJY5 A0A1A9XXZ7 A0A0C9RT37 A0A182IUW3 A0A1B0BX17 A0A182XXU8 A0A1A9WA71 A0A084VLE5 A0A1A9Z8J7 A0A1B0FIC8 A0A1A9UM59 B4M6Y8 A0A1Q3FUW7 A0A182TNA2 B3MS63 A0A154P3X4 W5JST5 A0A3B0JDW9 B4PZ92 A0A0M3QZE6 Q16UV6 A0A1S4FND0 B4JXI2 A0A151X588 A0A182XDT0 B3NXX0 Q9W3V1 A0A1W4UVS0 A0A1B6E7G7 A0A1B6DPY5 B4L512 A0A1B6MSA9 A0A0C5CUD5 B4MT40 Q29I77 A0A0P4VGW2 A0A1B6FFK2 T1I6U9 A0A1B6FU96 A0A2S2RBL2 F4WMI9 B4GWJ5 A0A0A9VRR7 A0A0K8TAJ9 X1WJN6 A0A146L7K0 A0A146LM60 A0A023EZV2 A0A3Q0IW64
Pubmed
19121390
26354079
28756777
26227816
28004739
25830018
+ More
26108605 23537049 24495485 25348373 24508170 30249741 12364791 14747013 17210077 26369729 24845553 28648823 20075255 20798317 21282665 25315136 21347285 25244985 24438588 17994087 20920257 23761445 17550304 17510324 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25224926 15632085 27129103 21719571 25401762 27192063 26823975 25474469
26108605 23537049 24495485 25348373 24508170 30249741 12364791 14747013 17210077 26369729 24845553 28648823 20075255 20798317 21282665 25315136 21347285 25244985 24438588 17994087 20920257 23761445 17550304 17510324 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25224926 15632085 27129103 21719571 25401762 27192063 26823975 25474469
EMBL
BABH01038624
ODYU01007763
SOQ50813.1
KQ460642
KPJ13351.1
KQ459592
+ More
KPI96921.1 KZ149931 PZC77329.1 NWSH01000292 PCG77687.1 JTDY01005326 KOB67109.1 GFDF01001538 JAV12546.1 GEZM01103172 JAV51492.1 LJIG01022620 KRT79591.1 GBXI01014073 JAD00219.1 JRES01001295 KNC23841.1 GDHF01032314 JAI20000.1 KB629993 ERL83318.1 APGK01055224 KB741261 ENN71614.1 GAMC01002508 JAC04048.1 KY426794 GAKP01012383 AQR60113.1 JAC46569.1 KU739357 ARA90534.1 NEVH01010480 PNF32392.1 KK107231 QOIP01000009 EZA54975.1 RLU18599.1 AAAB01008846 EAA06276.5 GDAI01002452 JAI15151.1 KK853123 KDR11066.1 AXCM01000448 NNAY01000889 OXU26053.1 AAZX01005110 KQ761155 OAD58156.1 LBMM01004181 KMQ92726.1 KZ288473 PBC25307.1 JR048928 AEY60805.1 GL437365 EFN70553.1 KQ435756 KOX75966.1 GL761846 EFZ22692.1 GL451188 EFN79802.1 CVRI01000020 CRK90600.1 KQ981954 KYN32173.1 KQ980629 KYN14993.1 AXCN02001239 ADTU01010268 ADTU01010269 ADTU01010270 KQ976600 KYM79475.1 KQ977642 KYN01040.1 GBYB01011830 JAG81597.1 JXJN01022027 ATLV01014483 KE524974 KFB38789.1 CCAG010018156 CH940653 EDW62555.1 GFDL01003671 JAV31374.1 CH902622 EDV34618.1 KQ434809 KZC06547.1 ADMH02000572 ETN65824.1 OUUW01000003 SPP78843.1 CM000162 EDX01059.2 CP012528 ALC49239.1 CH477613 EAT38324.1 CH916376 EDV95458.1 KQ982519 KYQ55484.1 CH954180 EDV47421.1 KQS30508.1 AE014298 AY118627 AAF46212.1 AAM49996.1 GEDC01003416 JAS33882.1 GEDC01009571 JAS27727.1 CH933811 EDW06271.1 GEBQ01001163 JAT38814.1 KM217110 AJO25037.1 CH963851 EDW75279.2 CH379063 EAL32776.1 GDKW01003007 JAI53588.1 GECZ01020783 GECZ01004445 JAS48986.1 JAS65324.1 ACPB03010146 GECZ01015973 GECZ01011686 JAS53796.1 JAS58083.1 GGMS01017957 MBY87160.1 GL888217 EGI64600.1 CH479194 EDW27079.1 KT717332 GBHO01044532 GBHO01013142 GBHO01007575 GBHO01007544 GBHO01007509 GBHO01003362 GBHO01003360 ANC95006.1 JAF99071.1 JAG30462.1 JAG36029.1 JAG36060.1 JAG36095.1 JAG40242.1 JAG40244.1 GBRD01003267 GBRD01000243 JAG62554.1 ABLF02037631 GDHC01015989 JAQ02640.1 GDHC01016291 GDHC01009515 GDHC01005383 JAQ02338.1 JAQ09114.1 JAQ13246.1 GBBI01003937 JAC14775.1
KPI96921.1 KZ149931 PZC77329.1 NWSH01000292 PCG77687.1 JTDY01005326 KOB67109.1 GFDF01001538 JAV12546.1 GEZM01103172 JAV51492.1 LJIG01022620 KRT79591.1 GBXI01014073 JAD00219.1 JRES01001295 KNC23841.1 GDHF01032314 JAI20000.1 KB629993 ERL83318.1 APGK01055224 KB741261 ENN71614.1 GAMC01002508 JAC04048.1 KY426794 GAKP01012383 AQR60113.1 JAC46569.1 KU739357 ARA90534.1 NEVH01010480 PNF32392.1 KK107231 QOIP01000009 EZA54975.1 RLU18599.1 AAAB01008846 EAA06276.5 GDAI01002452 JAI15151.1 KK853123 KDR11066.1 AXCM01000448 NNAY01000889 OXU26053.1 AAZX01005110 KQ761155 OAD58156.1 LBMM01004181 KMQ92726.1 KZ288473 PBC25307.1 JR048928 AEY60805.1 GL437365 EFN70553.1 KQ435756 KOX75966.1 GL761846 EFZ22692.1 GL451188 EFN79802.1 CVRI01000020 CRK90600.1 KQ981954 KYN32173.1 KQ980629 KYN14993.1 AXCN02001239 ADTU01010268 ADTU01010269 ADTU01010270 KQ976600 KYM79475.1 KQ977642 KYN01040.1 GBYB01011830 JAG81597.1 JXJN01022027 ATLV01014483 KE524974 KFB38789.1 CCAG010018156 CH940653 EDW62555.1 GFDL01003671 JAV31374.1 CH902622 EDV34618.1 KQ434809 KZC06547.1 ADMH02000572 ETN65824.1 OUUW01000003 SPP78843.1 CM000162 EDX01059.2 CP012528 ALC49239.1 CH477613 EAT38324.1 CH916376 EDV95458.1 KQ982519 KYQ55484.1 CH954180 EDV47421.1 KQS30508.1 AE014298 AY118627 AAF46212.1 AAM49996.1 GEDC01003416 JAS33882.1 GEDC01009571 JAS27727.1 CH933811 EDW06271.1 GEBQ01001163 JAT38814.1 KM217110 AJO25037.1 CH963851 EDW75279.2 CH379063 EAL32776.1 GDKW01003007 JAI53588.1 GECZ01020783 GECZ01004445 JAS48986.1 JAS65324.1 ACPB03010146 GECZ01015973 GECZ01011686 JAS53796.1 JAS58083.1 GGMS01017957 MBY87160.1 GL888217 EGI64600.1 CH479194 EDW27079.1 KT717332 GBHO01044532 GBHO01013142 GBHO01007575 GBHO01007544 GBHO01007509 GBHO01003362 GBHO01003360 ANC95006.1 JAF99071.1 JAG30462.1 JAG36029.1 JAG36060.1 JAG36095.1 JAG40242.1 JAG40244.1 GBRD01003267 GBRD01000243 JAG62554.1 ABLF02037631 GDHC01015989 JAQ02640.1 GDHC01016291 GDHC01009515 GDHC01005383 JAQ02338.1 JAQ09114.1 JAQ13246.1 GBBI01003937 JAC14775.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000037510
UP000037069
+ More
UP000030742 UP000019118 UP000235965 UP000095300 UP000075903 UP000053097 UP000279307 UP000007062 UP000075920 UP000027135 UP000075883 UP000215335 UP000002358 UP000075884 UP000036403 UP000242457 UP000000311 UP000005203 UP000075901 UP000053105 UP000008237 UP000183832 UP000069272 UP000095301 UP000078541 UP000075885 UP000078492 UP000075886 UP000005205 UP000078540 UP000078542 UP000092443 UP000075880 UP000092460 UP000076408 UP000091820 UP000030765 UP000092445 UP000092444 UP000078200 UP000008792 UP000075902 UP000007801 UP000076502 UP000000673 UP000268350 UP000002282 UP000092553 UP000008820 UP000001070 UP000075809 UP000076407 UP000008711 UP000000803 UP000192221 UP000009192 UP000007798 UP000001819 UP000015103 UP000007755 UP000008744 UP000007819 UP000079169
UP000030742 UP000019118 UP000235965 UP000095300 UP000075903 UP000053097 UP000279307 UP000007062 UP000075920 UP000027135 UP000075883 UP000215335 UP000002358 UP000075884 UP000036403 UP000242457 UP000000311 UP000005203 UP000075901 UP000053105 UP000008237 UP000183832 UP000069272 UP000095301 UP000078541 UP000075885 UP000078492 UP000075886 UP000005205 UP000078540 UP000078542 UP000092443 UP000075880 UP000092460 UP000076408 UP000091820 UP000030765 UP000092445 UP000092444 UP000078200 UP000008792 UP000075902 UP000007801 UP000076502 UP000000673 UP000268350 UP000002282 UP000092553 UP000008820 UP000001070 UP000075809 UP000076407 UP000008711 UP000000803 UP000192221 UP000009192 UP000007798 UP000001819 UP000015103 UP000007755 UP000008744 UP000007819 UP000079169
Interpro
Gene 3D
ProteinModelPortal
H9J8L7
A0A2H1WCM1
A0A0N1IFU3
A0A194PU30
A0A2W1BQG7
A0A2A4K0C3
+ More
A0A0L7KVP9 A0A1L8E1M5 A0A1Y1JQQ8 A0A0T6AYA6 A0A0A1WME4 A0A0L0BUY6 A0A0K8U0Z8 U4TTQ9 N6SVA0 W8BXL9 A0A034VVU5 A0A2P0P9G2 A0A2J7QUX9 A0A1I8NSV0 A0A182V6E0 A0A026WFT6 Q7QFE2 A0A0K8TLZ9 A0A182W5G7 A0A067QY08 A0A182M3W6 A0A232F6F5 K7IT18 A0A310SRX5 A0A182N1A3 A0A0J7KR08 A0A2A3E0P8 V9IIB8 E2A7N2 A0A088A858 A0A182T4N0 A0A0M9A3Y2 E9I9P7 E2BWT8 A0A1J1HR99 A0A182FJA1 A0A1I8MB77 A0A195EVA5 A0A182PBK0 A0A151J0Q1 A0A182QCP1 A0A158NAE7 A0A151I0U7 A0A195CJY5 A0A1A9XXZ7 A0A0C9RT37 A0A182IUW3 A0A1B0BX17 A0A182XXU8 A0A1A9WA71 A0A084VLE5 A0A1A9Z8J7 A0A1B0FIC8 A0A1A9UM59 B4M6Y8 A0A1Q3FUW7 A0A182TNA2 B3MS63 A0A154P3X4 W5JST5 A0A3B0JDW9 B4PZ92 A0A0M3QZE6 Q16UV6 A0A1S4FND0 B4JXI2 A0A151X588 A0A182XDT0 B3NXX0 Q9W3V1 A0A1W4UVS0 A0A1B6E7G7 A0A1B6DPY5 B4L512 A0A1B6MSA9 A0A0C5CUD5 B4MT40 Q29I77 A0A0P4VGW2 A0A1B6FFK2 T1I6U9 A0A1B6FU96 A0A2S2RBL2 F4WMI9 B4GWJ5 A0A0A9VRR7 A0A0K8TAJ9 X1WJN6 A0A146L7K0 A0A146LM60 A0A023EZV2 A0A3Q0IW64
A0A0L7KVP9 A0A1L8E1M5 A0A1Y1JQQ8 A0A0T6AYA6 A0A0A1WME4 A0A0L0BUY6 A0A0K8U0Z8 U4TTQ9 N6SVA0 W8BXL9 A0A034VVU5 A0A2P0P9G2 A0A2J7QUX9 A0A1I8NSV0 A0A182V6E0 A0A026WFT6 Q7QFE2 A0A0K8TLZ9 A0A182W5G7 A0A067QY08 A0A182M3W6 A0A232F6F5 K7IT18 A0A310SRX5 A0A182N1A3 A0A0J7KR08 A0A2A3E0P8 V9IIB8 E2A7N2 A0A088A858 A0A182T4N0 A0A0M9A3Y2 E9I9P7 E2BWT8 A0A1J1HR99 A0A182FJA1 A0A1I8MB77 A0A195EVA5 A0A182PBK0 A0A151J0Q1 A0A182QCP1 A0A158NAE7 A0A151I0U7 A0A195CJY5 A0A1A9XXZ7 A0A0C9RT37 A0A182IUW3 A0A1B0BX17 A0A182XXU8 A0A1A9WA71 A0A084VLE5 A0A1A9Z8J7 A0A1B0FIC8 A0A1A9UM59 B4M6Y8 A0A1Q3FUW7 A0A182TNA2 B3MS63 A0A154P3X4 W5JST5 A0A3B0JDW9 B4PZ92 A0A0M3QZE6 Q16UV6 A0A1S4FND0 B4JXI2 A0A151X588 A0A182XDT0 B3NXX0 Q9W3V1 A0A1W4UVS0 A0A1B6E7G7 A0A1B6DPY5 B4L512 A0A1B6MSA9 A0A0C5CUD5 B4MT40 Q29I77 A0A0P4VGW2 A0A1B6FFK2 T1I6U9 A0A1B6FU96 A0A2S2RBL2 F4WMI9 B4GWJ5 A0A0A9VRR7 A0A0K8TAJ9 X1WJN6 A0A146L7K0 A0A146LM60 A0A023EZV2 A0A3Q0IW64
PDB
5WVH
E-value=7.8076e-63,
Score=610
Ontologies
PATHWAY
GO
Topology
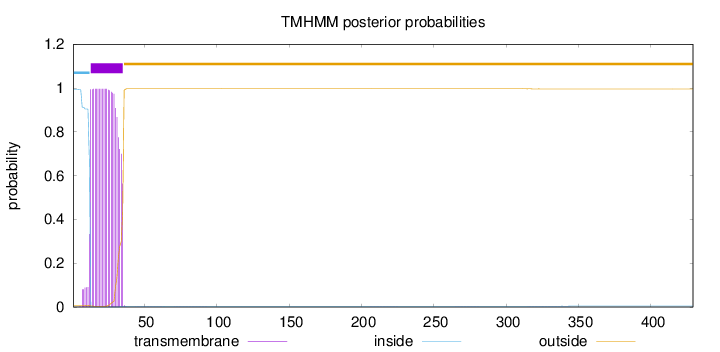
Length:
429
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.21409
Exp number, first 60 AAs:
22.18164
Total prob of N-in:
0.99339
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 35
outside
36 - 429
Population Genetic Test Statistics
Pi
0.549168
Theta
6.740461
Tajima's D
-2.613899
CLR
25.638406
CSRT
9.99950002499875e-05
Interpretation
Uncertain
