Gene
KWMTBOMO07557
Pre Gene Modal
BGIBMGA005849
Annotation
sid-1-related_gene3_precursor_[Bombyx_mori]
Full name
SID1 transmembrane family member 1
Location in the cell
PlasmaMembrane Reliability : 4.657
Sequence
CDS
ATGTTGTCCCGCGACCGCCTGCCATGTGTGTGTGTGTGCGCAGAGAAGTCGAGCCTGGCGGTGGCGGCGCGGGGCCTGCGGGCGCGGCCGCTGTACACGCCGCGGCTGGTCATGCTGCTCATCGCCAACGCCGCCAACTGGGGCTTCGCCATCTACGGGCTCCTGACCCATGCGGGGGACATAGCGACCCACCTGCTGAACGTGCTACTGTGCAACACGCTGCTGTACATCGTGTTCTACGTGCTGATGAAGCTGCTGCACGGGGAGCGCATCCGCTGGTACTCCTGGTGCTTCTTGGCCGCGGCGGCCGCCTGCTGGGTGCCAGCTCTCTACTTTTTTACTTCCGGCAGCACGGACTGGTCGGCGACGCCGGCGCGGTCCCGGCACCGCAACCACGAGTGCCGCGTGCTGCAGTTCTACGACTCGCACGACCTGTGGCACATGCTGTCGGCCGCCGCGCTCTACTTCACCTTCAACGTCATGCTCACCTGGGACGACGGACTCTCCGCCGTCAAACGGACAGAGATTGCCGTCTTTTGA
Protein
MLSRDRLPCVCVCAEKSSLAVAARGLRARPLYTPRLVMLLIANAANWGFAIYGLLTHAGDIATHLLNVLLCNTLLYIVFYVLMKLLHGERIRWYSWCFLAAAAACWVPALYFFTSGSTDWSATPARSRHRNHECRVLQFYDSHDLWHMLSAAALYFTFNVMLTWDDGLSAVKRTEIAVF
Summary
Description
In vitro binds long double-stranded RNA (dsRNA) (500 and 700 base pairs), but not dsRNA shorter than 300 bp. Not involved in RNA autophagy, a process in which RNA is directly imported into lysosomes in an ATP-dependent manner, and degraded.
Similarity
Belongs to the SID1 family.
Keywords
Alternative splicing
Complete proteome
Glycoprotein
Membrane
Polymorphism
Reference proteome
RNA-binding
Signal
Transmembrane
Transmembrane helix
Feature
chain SID1 transmembrane family member 1
splice variant In isoform 2.
sequence variant In dbSNP:rs9879313.
splice variant In isoform 2.
sequence variant In dbSNP:rs9879313.
Uniprot
A9CQL1
S5RRN7
H9J8K7
A0A2W1BSL8
A0A385JPJ4
A0A2A4K1B3
+ More
A0A0L7LM60 V9VMZ4 A0A212F6B3 A0A1E1W8F2 A0A194PVA2 A0A0N0PCB9 A0A2A4J7H1 A0A2A4IUB4 A0A2H1VGM1 V9VLL3 A0A1Y1KT41 A0A1Y1KTV8 B9W070 A0A1W4WGG4 A0A3S2M533 V4BXN2 D2A688 A7YFW2 A0A0N9JWV7 A9CQL3 A0A226ET70 A0A0N1I3U4 A0A059VQN8 A0A2J7RDI9 A0A194PSM1 A0A0S7K5X0 A0A067R6J9 A0A3Q2D4B8 A0A2J7RDJ2 A0A2J7RDJ6 A0A2J7RDK3 A0A3Q1JIW3 A0A3Q1JIT7 A0A3R7M6P4 A0A3B3ZUK6 A0A3Q1JPZ3 A0A1B6FZQ0 A0A2R2MI98 A0A2R2MIQ4 A0A3S2MCC8 A0A2G9QHI9 A0A3Q1JD10 A0A2R2MIB4 A0A1A8GFS2 A0A1A7YPN4 N6TBL6 U4U416 V9VIE0 K1RMY0 A0A2C9JQS9 A0A2C9JQS7 E2AJI6 A0A1D2M1A9 A0A059TD68 A0A154PER2 K7J510 A0A232EXE9 L5L359 A0A3M6TLD3 K9LY69 A0A3S2L0S6 A0A2B4SZM3 A0A2Y9QZ70 A0A0L8FH08 A0A2K6K5J0 A0A0L8FGV0 A0A2K6R5N7 A0A1S3W347 A0A2K6K5E3 A0A2K6R5P7 A0A2K5JXG8 A0A2K5JXG1 A0A1S2ZAG7 A0A2K5F514 A0A2K5MUU9 A7RYF1 A0A2K5MUX7 A0A2K5F536 F7FZS1 X2J861 A0A2J8W2K9 Q9NXL6-2 A0A2K5YEK1 A0A2K5MUV4 A0A2J8W2K3 Q9NXL6 A0A2K5YEK5 A0A2K5QBJ1 M3W303 A0A2I2V064 B4E0H9 A0A2K5QBI9 A0A337SST3 K9J2X7
A0A0L7LM60 V9VMZ4 A0A212F6B3 A0A1E1W8F2 A0A194PVA2 A0A0N0PCB9 A0A2A4J7H1 A0A2A4IUB4 A0A2H1VGM1 V9VLL3 A0A1Y1KT41 A0A1Y1KTV8 B9W070 A0A1W4WGG4 A0A3S2M533 V4BXN2 D2A688 A7YFW2 A0A0N9JWV7 A9CQL3 A0A226ET70 A0A0N1I3U4 A0A059VQN8 A0A2J7RDI9 A0A194PSM1 A0A0S7K5X0 A0A067R6J9 A0A3Q2D4B8 A0A2J7RDJ2 A0A2J7RDJ6 A0A2J7RDK3 A0A3Q1JIW3 A0A3Q1JIT7 A0A3R7M6P4 A0A3B3ZUK6 A0A3Q1JPZ3 A0A1B6FZQ0 A0A2R2MI98 A0A2R2MIQ4 A0A3S2MCC8 A0A2G9QHI9 A0A3Q1JD10 A0A2R2MIB4 A0A1A8GFS2 A0A1A7YPN4 N6TBL6 U4U416 V9VIE0 K1RMY0 A0A2C9JQS9 A0A2C9JQS7 E2AJI6 A0A1D2M1A9 A0A059TD68 A0A154PER2 K7J510 A0A232EXE9 L5L359 A0A3M6TLD3 K9LY69 A0A3S2L0S6 A0A2B4SZM3 A0A2Y9QZ70 A0A0L8FH08 A0A2K6K5J0 A0A0L8FGV0 A0A2K6R5N7 A0A1S3W347 A0A2K6K5E3 A0A2K6R5P7 A0A2K5JXG8 A0A2K5JXG1 A0A1S2ZAG7 A0A2K5F514 A0A2K5MUU9 A7RYF1 A0A2K5MUX7 A0A2K5F536 F7FZS1 X2J861 A0A2J8W2K9 Q9NXL6-2 A0A2K5YEK1 A0A2K5MUV4 A0A2J8W2K3 Q9NXL6 A0A2K5YEK5 A0A2K5QBJ1 M3W303 A0A2I2V064 B4E0H9 A0A2K5QBI9 A0A337SST3 K9J2X7
Pubmed
EMBL
AB327184
BAF95806.1
KC292008
AGS18762.1
BABH01038626
KZ149931
+ More
PZC77331.1 MG696730 AXZ00255.1 NWSH01000292 PCG77684.1 JTDY01000598 KOB76515.1 KF717090 AHC98015.1 AGBW02010072 OWR49249.1 GDQN01007801 JAT83253.1 KQ459592 KPI96918.1 KQ460642 KPJ13349.1 NWSH01002727 PCG67618.1 NWSH01007994 PCG62730.1 ODYU01002469 SOQ40003.1 KF717088 AHC98013.1 GEZM01078209 GEZM01078208 JAV62855.1 GEZM01078212 GEZM01078211 JAV62846.1 FJ619650 ACM47363.1 RSAL01000031 RVE51591.1 KB201890 ESO93839.1 KQ971346 EFA05674.2 EF688529 ABU63674.1 KR153285 ALG36907.1 AB327185 BAF95807.1 LNIX01000002 OXA60011.1 KQ458887 KPJ04577.1 KF561252 AHZ97882.1 NEVH01005288 PNF38903.1 KQ459593 KPI96307.1 GBYX01220090 GBYX01220088 GBYX01220086 GBYX01220080 GBYX01220079 GBYX01220078 JAO72599.1 KK852663 KDR19041.1 PNF38907.1 PNF38902.1 PNF38905.1 QCYY01001891 ROT74464.1 GECZ01014119 JAS55650.1 CM012449 RVE64360.1 KV986914 PIO14975.1 HAEC01002538 SBQ70615.1 HADX01010375 SBP32607.1 APGK01036719 APGK01036720 APGK01036721 KB740941 ENN77664.1 KB632001 ERL87797.1 KF717089 AHC98014.1 JH817099 EKC42950.1 GL440027 EFN66381.1 LJIJ01007384 ODM86750.1 KF192314 AHY18684.1 KQ434889 KZC10355.1 NNAY01001757 OXU23015.1 KB030337 ELK18174.1 RCHS01003423 RMX42091.1 JN676095 AFQ00936.1 RSAL01000287 RVE42946.1 LSMT01000007 PFX34022.1 KQ432038 KOF62934.1 KOF62933.1 DS469553 EDO43450.1 KJ135007 AHN55087.1 NDHI03003402 PNJ64015.1 AK000181 AC055740 AC112128 BC117222 PNJ64014.1 AANG04001815 AK303385 BAG64441.1 GABZ01004366 JAA49159.1
PZC77331.1 MG696730 AXZ00255.1 NWSH01000292 PCG77684.1 JTDY01000598 KOB76515.1 KF717090 AHC98015.1 AGBW02010072 OWR49249.1 GDQN01007801 JAT83253.1 KQ459592 KPI96918.1 KQ460642 KPJ13349.1 NWSH01002727 PCG67618.1 NWSH01007994 PCG62730.1 ODYU01002469 SOQ40003.1 KF717088 AHC98013.1 GEZM01078209 GEZM01078208 JAV62855.1 GEZM01078212 GEZM01078211 JAV62846.1 FJ619650 ACM47363.1 RSAL01000031 RVE51591.1 KB201890 ESO93839.1 KQ971346 EFA05674.2 EF688529 ABU63674.1 KR153285 ALG36907.1 AB327185 BAF95807.1 LNIX01000002 OXA60011.1 KQ458887 KPJ04577.1 KF561252 AHZ97882.1 NEVH01005288 PNF38903.1 KQ459593 KPI96307.1 GBYX01220090 GBYX01220088 GBYX01220086 GBYX01220080 GBYX01220079 GBYX01220078 JAO72599.1 KK852663 KDR19041.1 PNF38907.1 PNF38902.1 PNF38905.1 QCYY01001891 ROT74464.1 GECZ01014119 JAS55650.1 CM012449 RVE64360.1 KV986914 PIO14975.1 HAEC01002538 SBQ70615.1 HADX01010375 SBP32607.1 APGK01036719 APGK01036720 APGK01036721 KB740941 ENN77664.1 KB632001 ERL87797.1 KF717089 AHC98014.1 JH817099 EKC42950.1 GL440027 EFN66381.1 LJIJ01007384 ODM86750.1 KF192314 AHY18684.1 KQ434889 KZC10355.1 NNAY01001757 OXU23015.1 KB030337 ELK18174.1 RCHS01003423 RMX42091.1 JN676095 AFQ00936.1 RSAL01000287 RVE42946.1 LSMT01000007 PFX34022.1 KQ432038 KOF62934.1 KOF62933.1 DS469553 EDO43450.1 KJ135007 AHN55087.1 NDHI03003402 PNJ64015.1 AK000181 AC055740 AC112128 BC117222 PNJ64014.1 AANG04001815 AK303385 BAG64441.1 GABZ01004366 JAA49159.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000007151
UP000053268
UP000053240
+ More
UP000192223 UP000283053 UP000030746 UP000007266 UP000198287 UP000235965 UP000027135 UP000265020 UP000265040 UP000283509 UP000261520 UP000085678 UP000019118 UP000030742 UP000005408 UP000076420 UP000000311 UP000094527 UP000076502 UP000002358 UP000215335 UP000010552 UP000275408 UP000225706 UP000248480 UP000053454 UP000233180 UP000233200 UP000079721 UP000233080 UP000233020 UP000233060 UP000001593 UP000002279 UP000005640 UP000233140 UP000233040 UP000011712
UP000192223 UP000283053 UP000030746 UP000007266 UP000198287 UP000235965 UP000027135 UP000265020 UP000265040 UP000283509 UP000261520 UP000085678 UP000019118 UP000030742 UP000005408 UP000076420 UP000000311 UP000094527 UP000076502 UP000002358 UP000215335 UP000010552 UP000275408 UP000225706 UP000248480 UP000053454 UP000233180 UP000233200 UP000079721 UP000233080 UP000233020 UP000233060 UP000001593 UP000002279 UP000005640 UP000233140 UP000233040 UP000011712
Interpro
SUPFAM
SSF50978
SSF50978
Gene 3D
ProteinModelPortal
A9CQL1
S5RRN7
H9J8K7
A0A2W1BSL8
A0A385JPJ4
A0A2A4K1B3
+ More
A0A0L7LM60 V9VMZ4 A0A212F6B3 A0A1E1W8F2 A0A194PVA2 A0A0N0PCB9 A0A2A4J7H1 A0A2A4IUB4 A0A2H1VGM1 V9VLL3 A0A1Y1KT41 A0A1Y1KTV8 B9W070 A0A1W4WGG4 A0A3S2M533 V4BXN2 D2A688 A7YFW2 A0A0N9JWV7 A9CQL3 A0A226ET70 A0A0N1I3U4 A0A059VQN8 A0A2J7RDI9 A0A194PSM1 A0A0S7K5X0 A0A067R6J9 A0A3Q2D4B8 A0A2J7RDJ2 A0A2J7RDJ6 A0A2J7RDK3 A0A3Q1JIW3 A0A3Q1JIT7 A0A3R7M6P4 A0A3B3ZUK6 A0A3Q1JPZ3 A0A1B6FZQ0 A0A2R2MI98 A0A2R2MIQ4 A0A3S2MCC8 A0A2G9QHI9 A0A3Q1JD10 A0A2R2MIB4 A0A1A8GFS2 A0A1A7YPN4 N6TBL6 U4U416 V9VIE0 K1RMY0 A0A2C9JQS9 A0A2C9JQS7 E2AJI6 A0A1D2M1A9 A0A059TD68 A0A154PER2 K7J510 A0A232EXE9 L5L359 A0A3M6TLD3 K9LY69 A0A3S2L0S6 A0A2B4SZM3 A0A2Y9QZ70 A0A0L8FH08 A0A2K6K5J0 A0A0L8FGV0 A0A2K6R5N7 A0A1S3W347 A0A2K6K5E3 A0A2K6R5P7 A0A2K5JXG8 A0A2K5JXG1 A0A1S2ZAG7 A0A2K5F514 A0A2K5MUU9 A7RYF1 A0A2K5MUX7 A0A2K5F536 F7FZS1 X2J861 A0A2J8W2K9 Q9NXL6-2 A0A2K5YEK1 A0A2K5MUV4 A0A2J8W2K3 Q9NXL6 A0A2K5YEK5 A0A2K5QBJ1 M3W303 A0A2I2V064 B4E0H9 A0A2K5QBI9 A0A337SST3 K9J2X7
A0A0L7LM60 V9VMZ4 A0A212F6B3 A0A1E1W8F2 A0A194PVA2 A0A0N0PCB9 A0A2A4J7H1 A0A2A4IUB4 A0A2H1VGM1 V9VLL3 A0A1Y1KT41 A0A1Y1KTV8 B9W070 A0A1W4WGG4 A0A3S2M533 V4BXN2 D2A688 A7YFW2 A0A0N9JWV7 A9CQL3 A0A226ET70 A0A0N1I3U4 A0A059VQN8 A0A2J7RDI9 A0A194PSM1 A0A0S7K5X0 A0A067R6J9 A0A3Q2D4B8 A0A2J7RDJ2 A0A2J7RDJ6 A0A2J7RDK3 A0A3Q1JIW3 A0A3Q1JIT7 A0A3R7M6P4 A0A3B3ZUK6 A0A3Q1JPZ3 A0A1B6FZQ0 A0A2R2MI98 A0A2R2MIQ4 A0A3S2MCC8 A0A2G9QHI9 A0A3Q1JD10 A0A2R2MIB4 A0A1A8GFS2 A0A1A7YPN4 N6TBL6 U4U416 V9VIE0 K1RMY0 A0A2C9JQS9 A0A2C9JQS7 E2AJI6 A0A1D2M1A9 A0A059TD68 A0A154PER2 K7J510 A0A232EXE9 L5L359 A0A3M6TLD3 K9LY69 A0A3S2L0S6 A0A2B4SZM3 A0A2Y9QZ70 A0A0L8FH08 A0A2K6K5J0 A0A0L8FGV0 A0A2K6R5N7 A0A1S3W347 A0A2K6K5E3 A0A2K6R5P7 A0A2K5JXG8 A0A2K5JXG1 A0A1S2ZAG7 A0A2K5F514 A0A2K5MUU9 A7RYF1 A0A2K5MUX7 A0A2K5F536 F7FZS1 X2J861 A0A2J8W2K9 Q9NXL6-2 A0A2K5YEK1 A0A2K5MUV4 A0A2J8W2K3 Q9NXL6 A0A2K5YEK5 A0A2K5QBJ1 M3W303 A0A2I2V064 B4E0H9 A0A2K5QBI9 A0A337SST3 K9J2X7
Ontologies
GO
PANTHER
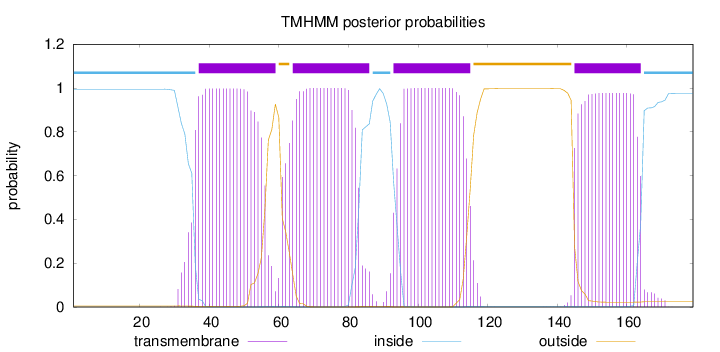
Topology
Subcellular location
Membrane
Length:
179
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
83.51569
Exp number, first 60 AAs:
21.45365
Total prob of N-in:
0.99492
POSSIBLE N-term signal
sequence
inside
1 - 36
TMhelix
37 - 59
outside
60 - 63
TMhelix
64 - 86
inside
87 - 92
TMhelix
93 - 115
outside
116 - 144
TMhelix
145 - 164
inside
165 - 179
Population Genetic Test Statistics
Pi
0.696075
Theta
4.171305
Tajima's D
-2.207315
CLR
212.063544
CSRT
0.00389980500974951
Interpretation
Possibly Positive selection
