Pre Gene Modal
BGIBMGA005845
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_serine_palmitoyltransferase_2_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.457 PlasmaMembrane Reliability : 1.056
Sequence
CDS
ATGAAGTCCACGCAGTCTAATGGATCAACGCCGAAGATGACACAAGTGGAGGGAGGCGGCCTGCCAAAGATAGACTGGTCCAAGTACGACACGTTCCCGGGCTCGTTCGAGAAGTGTTCCCTACTGACCGCCGTCCTCACGCACGTTGGACTTTACGTTCTCATGTTCCTGGGCTTCGTCAACCAGCTCATATTCAAGCCTAAGGCCGCCACCGAGAAGAACAGAGAGGGGTACGCTCCGCTGTACGATCCGTTCGAGCAGTTCTTCTCTCGGTACGTGTACCGGCGCGTGCGGCACTGCTTCAACCGACCGATATGCTCGGCGCCTGGAGCCGAAGTCGTCCTCAAGGAGTGGGAAAGCACCGATCACAATTGGACTTTCAAGTTCACGGGCGTGGAGCGGCGCTGCGTCAACCTGGGCTCGTACAACTACCTGGGGTTCGCGGGCGGGGAGGGCGCGGAGGGGGGCTGCGCGCGCGCCGTGGCCGACGCCGTGCGCCGCTACGGCATGGCGCAGGGCTCCTCGCGCGCCGAGCTCGGCACCACGCCGCTGCACCTCGAACTGGAACGGGCCGTGGCTGACTTCTTGGGGGTCGAGGACGCCATCGTGTTCGGAATGGGGTTCGCCACCAACACGCTGAGCATACCGGGGCTGATCGGGCCGGGCACGCTGGTGCTGAGCGACGAGAACAACCACGCGTCGCTCATCCTGGGCCTGCGCCTGTCCCGCGCCACCGTGCGCGTGTTCCGACACAACGACGTGCGCCACGCCGAGGAGCTGGCCCGCGCCGCGCTGGCCGAGCGCCGCTGGGCCAACGTGCTGCTGGTGGTGGAGGGCGTTTACAGCATGGAGGGCTCTGTGGCGGCACTGGAGGGCCTGGTGGCGTTGAAGCAGGCGCTGGGGCTGCACCTGTACGTGGACGAGGCGCACTCCGTGGGCGCTCTGGGGTCCACGGGCCGCGGCGCGCTGCAGCGGTGCCGCGTGCTCGCCGCGCACGTCGACGTGCTCATGGGCACCTTCACAAAGAGCTTCGGCGCGGCCGGGGGGTACATCGCAGGCCCGCGGCGGCTGGTGCAGTGGCTGCGCGCGCACGGGCACGCGCACGGCTACGCGCACAGCATGTCGTGCGGCGTGGCGGCGCAGGTGCTGGCGGCCGTGCGCGCGCTGGACGCGGCGCCGGGCCGGGCGCGGGTGCGGGCGCTGGCGCAGAACACGGCCTACTTCCGCCGCAGGCTGCGCGAGCTGGGCGTGGTGACGTTCGGGCACGACGACTCGCCGGTGGTGCCCATGCTGGTGTACACGTTCAGCAAGATGGCGGCCACGGTGGAGCGCCTCACGCGCGCCGGCGTCGCCACCGTCGGCGTCGGCTTCCCCGCCACGCCGCTCAACATGGCGCGCATACGTTTCTGCCTGTCGGCGTCCCACACCCGGGAGCAGCTGGACCGGTGCCTGCTGGCCGTGGAGCAGGTGGCGGACGAGCTGGGCCTGCGCTACTCGCGCCGCCGCCCCCTCGCGGAGCCCCGACCCTCGTAA
Protein
MKSTQSNGSTPKMTQVEGGGLPKIDWSKYDTFPGSFEKCSLLTAVLTHVGLYVLMFLGFVNQLIFKPKAATEKNREGYAPLYDPFEQFFSRYVYRRVRHCFNRPICSAPGAEVVLKEWESTDHNWTFKFTGVERRCVNLGSYNYLGFAGGEGAEGGCARAVADAVRRYGMAQGSSRAELGTTPLHLELERAVADFLGVEDAIVFGMGFATNTLSIPGLIGPGTLVLSDENNHASLILGLRLSRATVRVFRHNDVRHAEELARAALAERRWANVLLVVEGVYSMEGSVAALEGLVALKQALGLHLYVDEAHSVGALGSTGRGALQRCRVLAAHVDVLMGTFTKSFGAAGGYIAGPRRLVQWLRAHGHAHGYAHSMSCGVAAQVLAAVRALDAAPGRARVRALAQNTAYFRRRLRELGVVTFGHDDSPVVPMLVYTFSKMAATVERLTRAGVATVGVGFPATPLNMARIRFCLSASHTREQLDRCLLAVEQVADELGLRYSRRRPLAEPRPS
Summary
Cofactor
pyridoxal 5'-phosphate
Similarity
Belongs to the class-II pyridoxal-phosphate-dependent aminotransferase family.
Uniprot
A0A2A4K1P8
A0A2W1BZS8
A0A212F698
A0A158NY18
A0A195B8H6
A0A151IP31
+ More
A0A026VZ82 A0A0M9AA63 A0A182RBK6 A0A182Q3Y9 A0A182WT69 A0A182K1L1 A0A182V261 A0A151XBX8 A0A182W9P1 Q7Q3L8 A0A182HV64 A0A182L221 B0WCF6 A0A182YGT1 A0A1Q3F4Y2 A0A0A1WS35 A0A182MMX8 A0A1Q3F4Y8 A0A1Q3F4V7 Q17NH0 A0A034VLY0 A0A2M4ABT3 A0A195ED96 A0A0K8TSM6 E0VJB0 A0A1I8P7C6 A0A0C9RPX1 U5ETZ0 A0A087ZUZ3 A0A2A3E7E8 A0A1I8P7D0 A0A1I8P7H7 A0A0L7QRX2 A0A3L8D794 T1PIW1 A0A195FVQ6 A0A1B0GFA4 A0A067QVU3 A0A1A9YHI8 A0A1B0BKV9 A0A1A9Z8S9 A0A1J1HJE5 A0A1A9UT64 A0A1C9HJI7 B4MZ05 A0A3B0JNA3 A0A1W4VXB6 A0A1B0CZA0 A0A182F4K2 A0A1A9WUG2 B4NYA7 B4KLH6 B4HXQ8 B4LV60 A0A0R3P097 A0A0K8VNZ7 A0A2M4CZ25 B4Q6F5 B3N1C4 A0A0A9YLX6 B3N5Z3 Q9U912 Q9V3F2 D2A127 T1GSU6 Q29KV1 A0A0L0BRZ4 A0A139WJA7 B4GSS3 A0A1B6MR50 B4JQU7 E2BWP3 A0A1W4X4X2 A0A0V0G875 A0A1B6ERD8 A0A069DV97 A0A023F3S2 A0A1B6D108 A0A336KAB7 W5JH40 K7IQJ6 A0A2J7QIG8 A0A1L8DIN3 A0A224XHP9 A0A2R7W4H8 A0A0P4VWX8 A0A1Y1L908 A0A232FF07 A0A182HDR6 V5G047 A0A154PPN6 A0A0J7LAU3 T1I3Z1
A0A026VZ82 A0A0M9AA63 A0A182RBK6 A0A182Q3Y9 A0A182WT69 A0A182K1L1 A0A182V261 A0A151XBX8 A0A182W9P1 Q7Q3L8 A0A182HV64 A0A182L221 B0WCF6 A0A182YGT1 A0A1Q3F4Y2 A0A0A1WS35 A0A182MMX8 A0A1Q3F4Y8 A0A1Q3F4V7 Q17NH0 A0A034VLY0 A0A2M4ABT3 A0A195ED96 A0A0K8TSM6 E0VJB0 A0A1I8P7C6 A0A0C9RPX1 U5ETZ0 A0A087ZUZ3 A0A2A3E7E8 A0A1I8P7D0 A0A1I8P7H7 A0A0L7QRX2 A0A3L8D794 T1PIW1 A0A195FVQ6 A0A1B0GFA4 A0A067QVU3 A0A1A9YHI8 A0A1B0BKV9 A0A1A9Z8S9 A0A1J1HJE5 A0A1A9UT64 A0A1C9HJI7 B4MZ05 A0A3B0JNA3 A0A1W4VXB6 A0A1B0CZA0 A0A182F4K2 A0A1A9WUG2 B4NYA7 B4KLH6 B4HXQ8 B4LV60 A0A0R3P097 A0A0K8VNZ7 A0A2M4CZ25 B4Q6F5 B3N1C4 A0A0A9YLX6 B3N5Z3 Q9U912 Q9V3F2 D2A127 T1GSU6 Q29KV1 A0A0L0BRZ4 A0A139WJA7 B4GSS3 A0A1B6MR50 B4JQU7 E2BWP3 A0A1W4X4X2 A0A0V0G875 A0A1B6ERD8 A0A069DV97 A0A023F3S2 A0A1B6D108 A0A336KAB7 W5JH40 K7IQJ6 A0A2J7QIG8 A0A1L8DIN3 A0A224XHP9 A0A2R7W4H8 A0A0P4VWX8 A0A1Y1L908 A0A232FF07 A0A182HDR6 V5G047 A0A154PPN6 A0A0J7LAU3 T1I3Z1
Pubmed
28756777
22118469
21347285
24508170
12364791
14747013
+ More
17210077 20966253 25244985 25830018 17510324 25348373 26369729 20566863 30249741 25315136 24845553 17994087 17550304 15632085 22936249 25401762 26823975 10490662 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 26108605 20798317 26334808 25474469 20920257 23761445 20075255 27129103 28004739 28648823 26483478
17210077 20966253 25244985 25830018 17510324 25348373 26369729 20566863 30249741 25315136 24845553 17994087 17550304 15632085 22936249 25401762 26823975 10490662 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 26108605 20798317 26334808 25474469 20920257 23761445 20075255 27129103 28004739 28648823 26483478
EMBL
NWSH01000292
PCG77683.1
KZ149931
PZC77333.1
AGBW02010072
OWR49248.1
+ More
ADTU01002896 ADTU01002897 KQ976565 KYM80545.1 KQ976892 KYN07232.1 KK107558 EZA48985.1 KQ435701 KOX80375.1 AXCN02000864 KQ982316 KYQ57849.1 AAAB01008964 EAA12895.4 APCN01002494 DS231887 EDS43480.1 GFDL01012430 JAV22615.1 GBXI01012423 JAD01869.1 AXCM01005688 GFDL01012438 JAV22607.1 GFDL01012443 JAV22602.1 CH477199 EAT48248.1 GAKP01016157 JAC42795.1 GGFK01004747 MBW38068.1 KQ979074 KYN22792.1 GDAI01000224 JAI17379.1 DS235221 EEB13466.1 GBYB01010545 JAG80312.1 GANO01002566 JAB57305.1 KZ288345 PBC27635.1 KQ414766 KOC61385.1 QOIP01000012 RLU16159.1 KA647858 AFP62487.1 KQ981215 KYN44506.1 CCAG010012796 KK852886 KDR14360.1 JXJN01016056 CVRI01000001 CRK86590.1 KU737582 AOO86839.1 CH963913 EDW77344.1 OUUW01000006 SPP81832.1 AJVK01002205 AJVK01002206 CM000157 EDW89743.1 CH933807 EDW12857.1 CH480818 EDW51838.1 CH940649 EDW64320.1 CH379061 KRT04704.1 GDHF01027571 GDHF01011736 JAI24743.1 JAI40578.1 GGFL01006406 MBW70584.1 CM000361 CM002910 EDX05137.1 KMY90380.1 CH902652 EDV33645.2 GBHO01013054 GBRD01014680 GDHC01006864 JAG30550.1 JAG51146.1 JAQ11765.1 CH954177 EDV58031.1 AB017359 BAA83721.1 AE014134 AY095191 AAF53465.1 AAM12284.1 AHN54471.1 KQ971338 EFA02596.1 CAQQ02393656 CAQQ02393657 EAL33073.1 JRES01001454 KNC22812.1 KYB28108.1 CH479189 EDW25432.1 GEBQ01001591 JAT38386.1 CH916372 EDV99277.1 GL451165 EFN79898.1 GECL01001910 JAP04214.1 GECZ01029277 JAS40492.1 GBGD01001054 JAC87835.1 GBBI01002652 JAC16060.1 GEDC01018023 JAS19275.1 UFQS01000215 UFQT01000215 SSX01441.1 SSX21821.1 ADMH02001514 ETN62230.1 NEVH01013595 PNF28381.1 GFDF01007793 JAV06291.1 GFTR01007098 JAW09328.1 KK854321 PTY14633.1 GDKW01002502 JAI54093.1 GEZM01063137 JAV69281.1 NNAY01000360 OXU28897.1 JXUM01034762 JXUM01034763 KQ561035 KXJ79952.1 GALX01005136 JAB63330.1 KQ435007 KZC13707.1 LBMM01000102 KMR04938.1 ACPB03003297
ADTU01002896 ADTU01002897 KQ976565 KYM80545.1 KQ976892 KYN07232.1 KK107558 EZA48985.1 KQ435701 KOX80375.1 AXCN02000864 KQ982316 KYQ57849.1 AAAB01008964 EAA12895.4 APCN01002494 DS231887 EDS43480.1 GFDL01012430 JAV22615.1 GBXI01012423 JAD01869.1 AXCM01005688 GFDL01012438 JAV22607.1 GFDL01012443 JAV22602.1 CH477199 EAT48248.1 GAKP01016157 JAC42795.1 GGFK01004747 MBW38068.1 KQ979074 KYN22792.1 GDAI01000224 JAI17379.1 DS235221 EEB13466.1 GBYB01010545 JAG80312.1 GANO01002566 JAB57305.1 KZ288345 PBC27635.1 KQ414766 KOC61385.1 QOIP01000012 RLU16159.1 KA647858 AFP62487.1 KQ981215 KYN44506.1 CCAG010012796 KK852886 KDR14360.1 JXJN01016056 CVRI01000001 CRK86590.1 KU737582 AOO86839.1 CH963913 EDW77344.1 OUUW01000006 SPP81832.1 AJVK01002205 AJVK01002206 CM000157 EDW89743.1 CH933807 EDW12857.1 CH480818 EDW51838.1 CH940649 EDW64320.1 CH379061 KRT04704.1 GDHF01027571 GDHF01011736 JAI24743.1 JAI40578.1 GGFL01006406 MBW70584.1 CM000361 CM002910 EDX05137.1 KMY90380.1 CH902652 EDV33645.2 GBHO01013054 GBRD01014680 GDHC01006864 JAG30550.1 JAG51146.1 JAQ11765.1 CH954177 EDV58031.1 AB017359 BAA83721.1 AE014134 AY095191 AAF53465.1 AAM12284.1 AHN54471.1 KQ971338 EFA02596.1 CAQQ02393656 CAQQ02393657 EAL33073.1 JRES01001454 KNC22812.1 KYB28108.1 CH479189 EDW25432.1 GEBQ01001591 JAT38386.1 CH916372 EDV99277.1 GL451165 EFN79898.1 GECL01001910 JAP04214.1 GECZ01029277 JAS40492.1 GBGD01001054 JAC87835.1 GBBI01002652 JAC16060.1 GEDC01018023 JAS19275.1 UFQS01000215 UFQT01000215 SSX01441.1 SSX21821.1 ADMH02001514 ETN62230.1 NEVH01013595 PNF28381.1 GFDF01007793 JAV06291.1 GFTR01007098 JAW09328.1 KK854321 PTY14633.1 GDKW01002502 JAI54093.1 GEZM01063137 JAV69281.1 NNAY01000360 OXU28897.1 JXUM01034762 JXUM01034763 KQ561035 KXJ79952.1 GALX01005136 JAB63330.1 KQ435007 KZC13707.1 LBMM01000102 KMR04938.1 ACPB03003297
Proteomes
UP000218220
UP000007151
UP000005205
UP000078540
UP000078542
UP000053097
+ More
UP000053105 UP000075900 UP000075886 UP000076407 UP000075881 UP000075903 UP000075809 UP000075920 UP000007062 UP000075840 UP000075882 UP000002320 UP000076408 UP000075883 UP000008820 UP000078492 UP000009046 UP000095300 UP000005203 UP000242457 UP000053825 UP000279307 UP000095301 UP000078541 UP000092444 UP000027135 UP000092443 UP000092460 UP000092445 UP000183832 UP000078200 UP000007798 UP000268350 UP000192221 UP000092462 UP000069272 UP000091820 UP000002282 UP000009192 UP000001292 UP000008792 UP000001819 UP000000304 UP000007801 UP000008711 UP000000803 UP000007266 UP000015102 UP000037069 UP000008744 UP000001070 UP000008237 UP000192223 UP000000673 UP000002358 UP000235965 UP000215335 UP000069940 UP000249989 UP000076502 UP000036403 UP000015103
UP000053105 UP000075900 UP000075886 UP000076407 UP000075881 UP000075903 UP000075809 UP000075920 UP000007062 UP000075840 UP000075882 UP000002320 UP000076408 UP000075883 UP000008820 UP000078492 UP000009046 UP000095300 UP000005203 UP000242457 UP000053825 UP000279307 UP000095301 UP000078541 UP000092444 UP000027135 UP000092443 UP000092460 UP000092445 UP000183832 UP000078200 UP000007798 UP000268350 UP000192221 UP000092462 UP000069272 UP000091820 UP000002282 UP000009192 UP000001292 UP000008792 UP000001819 UP000000304 UP000007801 UP000008711 UP000000803 UP000007266 UP000015102 UP000037069 UP000008744 UP000001070 UP000008237 UP000192223 UP000000673 UP000002358 UP000235965 UP000215335 UP000069940 UP000249989 UP000076502 UP000036403 UP000015103
Interpro
SUPFAM
SSF53383
SSF53383
Gene 3D
ProteinModelPortal
A0A2A4K1P8
A0A2W1BZS8
A0A212F698
A0A158NY18
A0A195B8H6
A0A151IP31
+ More
A0A026VZ82 A0A0M9AA63 A0A182RBK6 A0A182Q3Y9 A0A182WT69 A0A182K1L1 A0A182V261 A0A151XBX8 A0A182W9P1 Q7Q3L8 A0A182HV64 A0A182L221 B0WCF6 A0A182YGT1 A0A1Q3F4Y2 A0A0A1WS35 A0A182MMX8 A0A1Q3F4Y8 A0A1Q3F4V7 Q17NH0 A0A034VLY0 A0A2M4ABT3 A0A195ED96 A0A0K8TSM6 E0VJB0 A0A1I8P7C6 A0A0C9RPX1 U5ETZ0 A0A087ZUZ3 A0A2A3E7E8 A0A1I8P7D0 A0A1I8P7H7 A0A0L7QRX2 A0A3L8D794 T1PIW1 A0A195FVQ6 A0A1B0GFA4 A0A067QVU3 A0A1A9YHI8 A0A1B0BKV9 A0A1A9Z8S9 A0A1J1HJE5 A0A1A9UT64 A0A1C9HJI7 B4MZ05 A0A3B0JNA3 A0A1W4VXB6 A0A1B0CZA0 A0A182F4K2 A0A1A9WUG2 B4NYA7 B4KLH6 B4HXQ8 B4LV60 A0A0R3P097 A0A0K8VNZ7 A0A2M4CZ25 B4Q6F5 B3N1C4 A0A0A9YLX6 B3N5Z3 Q9U912 Q9V3F2 D2A127 T1GSU6 Q29KV1 A0A0L0BRZ4 A0A139WJA7 B4GSS3 A0A1B6MR50 B4JQU7 E2BWP3 A0A1W4X4X2 A0A0V0G875 A0A1B6ERD8 A0A069DV97 A0A023F3S2 A0A1B6D108 A0A336KAB7 W5JH40 K7IQJ6 A0A2J7QIG8 A0A1L8DIN3 A0A224XHP9 A0A2R7W4H8 A0A0P4VWX8 A0A1Y1L908 A0A232FF07 A0A182HDR6 V5G047 A0A154PPN6 A0A0J7LAU3 T1I3Z1
A0A026VZ82 A0A0M9AA63 A0A182RBK6 A0A182Q3Y9 A0A182WT69 A0A182K1L1 A0A182V261 A0A151XBX8 A0A182W9P1 Q7Q3L8 A0A182HV64 A0A182L221 B0WCF6 A0A182YGT1 A0A1Q3F4Y2 A0A0A1WS35 A0A182MMX8 A0A1Q3F4Y8 A0A1Q3F4V7 Q17NH0 A0A034VLY0 A0A2M4ABT3 A0A195ED96 A0A0K8TSM6 E0VJB0 A0A1I8P7C6 A0A0C9RPX1 U5ETZ0 A0A087ZUZ3 A0A2A3E7E8 A0A1I8P7D0 A0A1I8P7H7 A0A0L7QRX2 A0A3L8D794 T1PIW1 A0A195FVQ6 A0A1B0GFA4 A0A067QVU3 A0A1A9YHI8 A0A1B0BKV9 A0A1A9Z8S9 A0A1J1HJE5 A0A1A9UT64 A0A1C9HJI7 B4MZ05 A0A3B0JNA3 A0A1W4VXB6 A0A1B0CZA0 A0A182F4K2 A0A1A9WUG2 B4NYA7 B4KLH6 B4HXQ8 B4LV60 A0A0R3P097 A0A0K8VNZ7 A0A2M4CZ25 B4Q6F5 B3N1C4 A0A0A9YLX6 B3N5Z3 Q9U912 Q9V3F2 D2A127 T1GSU6 Q29KV1 A0A0L0BRZ4 A0A139WJA7 B4GSS3 A0A1B6MR50 B4JQU7 E2BWP3 A0A1W4X4X2 A0A0V0G875 A0A1B6ERD8 A0A069DV97 A0A023F3S2 A0A1B6D108 A0A336KAB7 W5JH40 K7IQJ6 A0A2J7QIG8 A0A1L8DIN3 A0A224XHP9 A0A2R7W4H8 A0A0P4VWX8 A0A1Y1L908 A0A232FF07 A0A182HDR6 V5G047 A0A154PPN6 A0A0J7LAU3 T1I3Z1
PDB
3TQX
E-value=1.05899e-43,
Score=446
Ontologies
KEGG
PATHWAY
GO
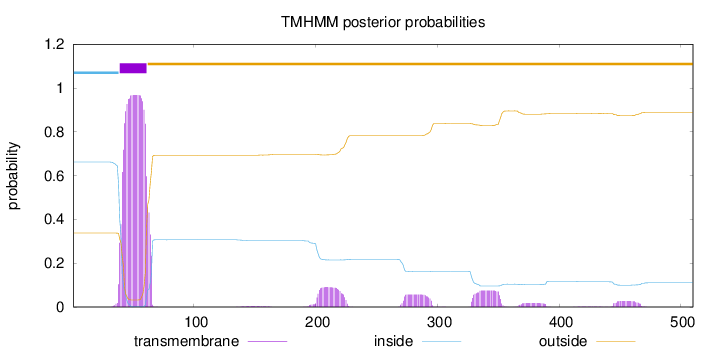
Topology
Length:
510
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
27.10976
Exp number, first 60 AAs:
19.04659
Total prob of N-in:
0.66206
POSSIBLE N-term signal
sequence
inside
1 - 38
TMhelix
39 - 61
outside
62 - 510
Population Genetic Test Statistics
Pi
1.293205
Theta
7.147803
Tajima's D
-2.403671
CLR
184.495986
CSRT
0.00109994500274986
Interpretation
Possibly Positive selection
