Gene
KWMTBOMO07554 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005860
Annotation
PREDICTED:_reticulon-3-like_isoform_X2_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.257
Sequence
CDS
ATGGAGAGCACAGGCGCCCTCCTGGGAGGCTTCGCCGCCGCCGGTGATGCGGCGCCCGGTGCGGATCAGACCATGAAGCGCGACCAGGACTCGACGGATGATTTCGAGCATCTCGATCGTGAAACTAAACAAGACCCGGCGGATTCACCGGTACACCATCATCGCGTGGCCACTCAGAGTTTCTTGGAGATGGAACGTGGGCCCGCAGCGGAACATCGGCCGCCTTCCGTCGCCGAAAAGCTTATGGACCACATGGCCGACAAATTTACCGACAGTGAGTCAGACGCCGATACTGCAGGAGAGTCGCCCCTCCATCGGCCCGAGCCGAGGGTCGAACTCCCGAAGCCTAGCGCTCCGACACCGGAACCACCGATGCCTGAAGCTCACGACCCGACACCGGTGCTGGCCCCCGCGCCCGCTCCGCCTGTCCCCGAGAAACCTGTGGAGCCCAAAGTGGAGCCGGCCGTCGTCCCTGAGCCTAAGCCGGTGCCGAAAGCGGAACCCGTCAAGCGAGAAGAGGTGGCTCCGCCCAAGCCTGAGCCAGCCTCGCAACCAAAGCCGGTCCCCGTAGAGGAGAAGCCGGAGCCGACTCGCGGACCTACGGCGCATGTCATCGAAGCAGAAGTAATCTTCTGTCAAATGGGACTTGTGGAGTCGTTGATCTACTGGCGGAACGCGGCACGCTCTGGCGCGGCCCTGGGCGCGGGGCTGGCGCTGCTGGTGGCGCTGGCGTGCTGCTCGGTGGTGTCGGTGCTCGCCTACGTGTCGCTGCTGGCGCTCAGCGCGGCCGTCGCCTTCCGCCTCTACAAGAACGTGCTGCAGGCCGTTCAGAAGACCAACGATGGACATCCCTTCAAATGGCTGCTGGAGGCGGAGGTGAGCGTGTCGGCGGAGCGCGCGCAGGCGCTGGCCGCCGCCGCCACCGCGCACCTCAACGCCGCCCTGTATGAGCTGCGCAGACTGTTCCTGGTGGAGGACCTGGTGGACTCGCTGAAGTTCGGCGTGCTGCTGTGGTGCCTCACGTACGTGGGGGCGTGCTTCAACGGGATCACGCTCATCATCCTCGGTTGGATAGCGCTGTTCACGCTGCCGAAGGCCTACGAGATGAACAAGGCGCAGGTGGACGCCAACCTCGAGCTGGCGCGCGCCAAGATCAACGAGATCACCGCCAAGGTGCGCGCCGCGGTGCCGCTGGGCAGGAGGGCGGAGGGCGACAAGGACAAGTAG
Protein
MESTGALLGGFAAAGDAAPGADQTMKRDQDSTDDFEHLDRETKQDPADSPVHHHRVATQSFLEMERGPAAEHRPPSVAEKLMDHMADKFTDSESDADTAGESPLHRPEPRVELPKPSAPTPEPPMPEAHDPTPVLAPAPAPPVPEKPVEPKVEPAVVPEPKPVPKAEPVKREEVAPPKPEPASQPKPVPVEEKPEPTRGPTAHVIEAEVIFCQMGLVESLIYWRNAARSGAALGAGLALLVALACCSVVSVLAYVSLLALSAAVAFRLYKNVLQAVQKTNDGHPFKWLLEAEVSVSAERAQALAAAATAHLNAALYELRRLFLVEDLVDSLKFGVLLWCLTYVGACFNGITLIILGWIALFTLPKAYEMNKAQVDANLELARAKINEITAKVRAAVPLGRRAEGDKDK
Summary
Uniprot
ProteinModelPortal
PDB
2KO2
E-value=1.11417e-10,
Score=160
Ontologies
GO
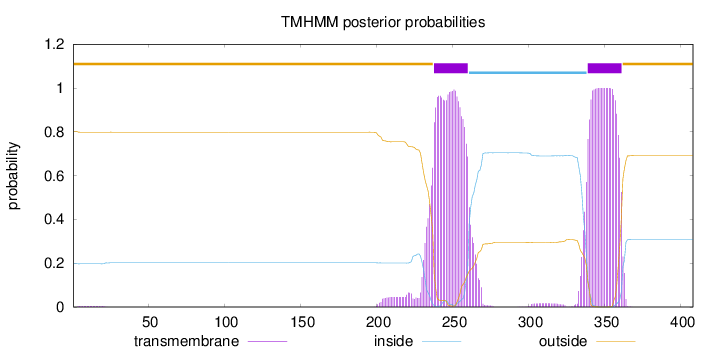
Topology
Length:
408
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
51.75702
Exp number, first 60 AAs:
0.08221
Total prob of N-in:
0.19727
outside
1 - 237
TMhelix
238 - 260
inside
261 - 338
TMhelix
339 - 361
outside
362 - 408
Population Genetic Test Statistics
Pi
21.770618
Theta
25.035679
Tajima's D
-2.336505
CLR
0.596243
CSRT
0.00279986000699965
Interpretation
Possibly Positive selection
