Gene
KWMTBOMO07551
Annotation
PREDICTED:_DNA-binding_protein_D-ETS-6-like_[Bombyx_mori]
Full name
Protein FEV
Alternative Name
PC12 ETS domain-containing transcription factor 1
Fifth Ewing variant protein
Fifth Ewing variant protein
Transcription factor
Location in the cell
Nuclear Reliability : 2.22
Sequence
CDS
ATGGCATCACGCTATGGGTATGCGGCTCTGAGTGCACGCGCTGGAGGAGGACAGGTGCAGCTCTGGCAGTTCCTGCTGGAGGAGCTGGCTGCAGGAGCTCCTGGAATCTGTTGGGAGGGTCCCCCTGGTGATGGAGAGTTCCGCCTGTCGGATCCGGATGAGGTGGCCCGGCGCTGGGGCCGGCGCAAACAGAAACCCAACATGAACTACGACAAACTATCGCGAGCGCTGCGGTACTATTATGATAAGAACATCATGAGCAAGGTACACGGGAAGCGCTACGCGTACAGGTTCAGTTGGGCGGGTCTGACCGCGGCGTGCCAGGCGCAGGCTCCGGACGCGCCCCCCTACTGGCACTACCTCCCGCCCCCCGGACACCCCCCGCAACATGTCCACGGGAACATCCACAGCTCGCAGCAGACCTAG
Protein
MASRYGYAALSARAGGGQVQLWQFLLEELAAGAPGICWEGPPGDGEFRLSDPDEVARRWGRRKQKPNMNYDKLSRALRYYYDKNIMSKVHGKRYAYRFSWAGLTAACQAQAPDAPPYWHYLPPPGHPPQHVHGNIHSSQQT
Summary
Description
Functions as a transcriptional regulator. May function as a transcriptional repressor. Functions in the differentiation and the maintenance of the central serotonergic neurons. May play a role in cell growth.
Functions as a transcriptional regulator. According to PubMed:12761502, it functions as a transcriptional repressor. Functions in the differentiation and the maintenance of the central serotonergic neurons. May play a role in cell growth.
Functions as a transcriptional regulator. According to PubMed:12761502, it functions as a transcriptional repressor. Functions in the differentiation and the maintenance of the central serotonergic neurons. May play a role in cell growth.
Similarity
Belongs to the ETS family.
Keywords
Complete proteome
Developmental protein
Differentiation
DNA-binding
Neurogenesis
Nucleus
Reference proteome
Transcription
Transcription regulation
3D-structure
Chromosomal rearrangement
Feature
chain Protein FEV
Uniprot
A0A2R8ZHN2
A0A341CW01
G3UMM1
A0A2K5Z6W4
A0A2K5HJU7
I3MXW7
+ More
A0A2U3YH87 F6WXL8 A0A0H2UHP3 G3WPL0 H0WCZ3 A0A384AL72 A0A0D9R8A4 A0A096N222 A0A2K5NMZ5 A0A2K6E7N4 A0A2K5X8V4 A0A2K6NPL9 O70132 Q8QZW2 A0A2Y9FUM8 A0A2K6N6R6 A0A2J8TUA6 A0A340WFE6 G3R5E2 A0A2I3RTZ5 Q99581 A0A2U4BMK6 A0A2R8MR30 W5QET0 A0A2K5RYE6 M3YYN9 A0A1U7QPQ1 A0A2K6GMR6
A0A2U3YH87 F6WXL8 A0A0H2UHP3 G3WPL0 H0WCZ3 A0A384AL72 A0A0D9R8A4 A0A096N222 A0A2K5NMZ5 A0A2K6E7N4 A0A2K5X8V4 A0A2K6NPL9 O70132 Q8QZW2 A0A2Y9FUM8 A0A2K6N6R6 A0A2J8TUA6 A0A340WFE6 G3R5E2 A0A2I3RTZ5 Q99581 A0A2U4BMK6 A0A2R8MR30 W5QET0 A0A2K5RYE6 M3YYN9 A0A1U7QPQ1 A0A2K6GMR6
Pubmed
EMBL
AJFE02063764
AJFE02063765
AGTP01062686
AGTP01062687
AC132020
AEFK01132713
+ More
AEFK01132714 AAKN02049531 AQIB01003880 AHZZ02004557 AQIA01015930 U91679 AY049085 AY049086 CH466548 BC138435 BC138436 NDHI03003483 PNJ36586.1 CABD030018694 AACZ04056567 NBAG03000230 PNI70823.1 Y08976 AC097468 CH471063 BC023511 AMGL01056043 AEYP01056834
AEFK01132714 AAKN02049531 AQIB01003880 AHZZ02004557 AQIA01015930 U91679 AY049085 AY049086 CH466548 BC138435 BC138436 NDHI03003483 PNJ36586.1 CABD030018694 AACZ04056567 NBAG03000230 PNI70823.1 Y08976 AC097468 CH471063 BC023511 AMGL01056043 AEYP01056834
Proteomes
UP000240080
UP000252040
UP000007646
UP000233140
UP000233080
UP000005215
+ More
UP000245341 UP000002280 UP000002494 UP000007648 UP000005447 UP000261681 UP000029965 UP000028761 UP000233060 UP000233120 UP000233100 UP000233200 UP000000589 UP000248484 UP000233180 UP000265300 UP000001519 UP000002277 UP000005640 UP000245320 UP000008225 UP000002356 UP000233040 UP000000715 UP000189706 UP000233160
UP000245341 UP000002280 UP000002494 UP000007648 UP000005447 UP000261681 UP000029965 UP000028761 UP000233060 UP000233120 UP000233100 UP000233200 UP000000589 UP000248484 UP000233180 UP000265300 UP000001519 UP000002277 UP000005640 UP000245320 UP000008225 UP000002356 UP000233040 UP000000715 UP000189706 UP000233160
Pfam
PF00178 Ets
SUPFAM
SSF46785
SSF46785
Gene 3D
ProteinModelPortal
A0A2R8ZHN2
A0A341CW01
G3UMM1
A0A2K5Z6W4
A0A2K5HJU7
I3MXW7
+ More
A0A2U3YH87 F6WXL8 A0A0H2UHP3 G3WPL0 H0WCZ3 A0A384AL72 A0A0D9R8A4 A0A096N222 A0A2K5NMZ5 A0A2K6E7N4 A0A2K5X8V4 A0A2K6NPL9 O70132 Q8QZW2 A0A2Y9FUM8 A0A2K6N6R6 A0A2J8TUA6 A0A340WFE6 G3R5E2 A0A2I3RTZ5 Q99581 A0A2U4BMK6 A0A2R8MR30 W5QET0 A0A2K5RYE6 M3YYN9 A0A1U7QPQ1 A0A2K6GMR6
A0A2U3YH87 F6WXL8 A0A0H2UHP3 G3WPL0 H0WCZ3 A0A384AL72 A0A0D9R8A4 A0A096N222 A0A2K5NMZ5 A0A2K6E7N4 A0A2K5X8V4 A0A2K6NPL9 O70132 Q8QZW2 A0A2Y9FUM8 A0A2K6N6R6 A0A2J8TUA6 A0A340WFE6 G3R5E2 A0A2I3RTZ5 Q99581 A0A2U4BMK6 A0A2R8MR30 W5QET0 A0A2K5RYE6 M3YYN9 A0A1U7QPQ1 A0A2K6GMR6
PDB
3ZP5
E-value=7.42028e-32,
Score=336
Ontologies
GO
GO:0005634
GO:0010628
GO:0043565
GO:0048665
GO:0042551
GO:0003700
GO:0016607
GO:0030154
GO:0000981
GO:0006357
GO:0007399
GO:0045944
GO:0003690
GO:0001228
GO:0000978
GO:0045893
GO:0051611
GO:0043025
GO:1905627
GO:0006366
GO:0003714
GO:0006355
GO:0004190
GO:0006508
GO:0015914
GO:0016021
GO:0016020
GO:0003707
GO:0051537
Topology
Subcellular location
Nucleus
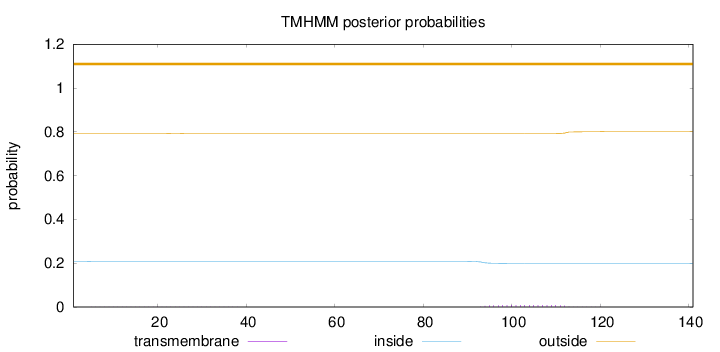
Length:
141
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.17639
Exp number, first 60 AAs:
0.01726
Total prob of N-in:
0.20835
outside
1 - 141
Population Genetic Test Statistics
Pi
20.68444
Theta
25.748273
Tajima's D
-0.602555
CLR
0.534124
CSRT
0.217089145542723
Interpretation
Uncertain
