Pre Gene Modal
BGIBMGA005864
Annotation
PREDICTED:_trafficking_protein_particle_complex_subunit_4_[Plutella_xylostella]
Location in the cell
Cytoplasmic Reliability : 1.058 PlasmaMembrane Reliability : 1.745
Sequence
CDS
ATGGTTATATACGGAATTTATATTGTTAGCAAGTCTGGTGGTCTCATCTACAACTACGATCACAATATTCCTCGAGTGGAGACTGAGAAAACGTTTGGATTTCCCCTTGATATTAAATTGCAGTATGAAAATAAAAAAGTTGTTGTGGCATTTGGCCAAAGAGATGGAATAAATGTTGGACATGTATTACTATCAGTAAATGGATCTCCAGTGACAGGAAGAACAACGGAAGACAATAGAGATGTCTTTGATATTATAGAAGCTAAGGAAAACTATCCTCTGAGTCTAAAATTTGGTAGAGTGAGAGCAACAACAAATGAGAAGATAGTGCTGGCCAGCATGTTCTATCCTCTTTTTGCCCTTGCTAGTCAGCTCAGTCCCATACCAAAATCCTCAGGAATAGAATCCTTAACGGCTGACACATTCAAGTTGTCGTGTTTTCAAACACTCACTGGTGTAAAATTTATAATAGTAACAGACACTAATATGCAAGGTACTGATGTGGTCCTAAAGAGGATCTATGAGTTATATTCAGACTATGCACTTAAGAATCCATTCTACTCACTCGAGATGCCAATAAGATGTGAACTGTTCGACACATCTCTACACACACTGCTGGAACTAGTTGAGAAGTCTGGGACTGCTAATTTATAA
Protein
MVIYGIYIVSKSGGLIYNYDHNIPRVETEKTFGFPLDIKLQYENKKVVVAFGQRDGINVGHVLLSVNGSPVTGRTTEDNRDVFDIIEAKENYPLSLKFGRVRATTNEKIVLASMFYPLFALASQLSPIPKSSGIESLTADTFKLSCFQTLTGVKFIIVTDTNMQGTDVVLKRIYELYSDYALKNPFYSLEMPIRCELFDTSLHTLLELVEKSGTANL
Summary
Uniprot
H9J8M2
A0A1E1W2S6
A0A2H1WZY7
A0A2W1BX02
A0A2A4JJE4
A0A212F677
+ More
A0A0N1IH47 S4P460 E3UPC9 A0A194PUA5 A0A2J7PK69 A0A0L7L5C2 A0A1Y1KRE6 A0A1W4X4U1 D2A131 A0A310SAE1 A0A0M9ACC3 A0A067R3S4 E2BWP0 E1ZZF1 A0A154PPB0 A0A026VYY9 A0A0L7QRY1 A0A087ZUZ4 A0A2A3E792 A0A232FG73 K7IR56 J3JVV9 A0A195ECF5 F4WNQ8 A0A195FVM0 A0A151IP55 Q7Q3Z0 A0A240PKQ6 A0A158NVP7 A0A1Y9HE26 A0A182GWG8 A0A084WGW4 A0A1L8DVQ4 A0A182Y069 A0A023EIU6 A0A240PMR3 A0A182WU42 A0A182L1H6 A0A182HUQ4 A0A2M3ZJV9 A0A0L0BUR8 A0A182UN56 A0A2M4BZ90 A0A1Y9H2Y4 A0A1Y9IW44 A0A1Y9G9G4 A0A2M4AQ60 A0A1Y9H9J4 A0A182UDH9 A0A2M4BZU3 W5JMH8 A0A182MIT9 A0A240PL21 A0A1I8N5Y8 U5EV61 A0A1B6D6Q7 A0A1Q3FDM4 A0A0K8TSD5 A0A1B6JFD7 B0X6Q2 A0A1B6FXS1 A0A1J1INN1 A0A1B6M0G6 A0A1I8QF50 A0A2S2Q9P7 B3N7D6 B4NXH0 B4HYQ3 B4Q786 E0VZF3 Q9VLI9 Q29JT8 B4N7T0 A0A0V0GAK5 A0A023EJV3 A0A1A9XEF1 A0A1B0AYT9 A0A3B0JL02 A0A1A9UYL4 A0A0P4VT81 T1HV27 A0A2S2PI94 G8JKE9 A0A069DQ86 B4JDZ6 B3MLY8 A0A2R7X2M3 B4LSE5 A0A2H8TPB8 A0A1W4W084 A0A0A1WTN3 A0A1B0DEK5 W8C8H1 A0A146KSS1
A0A0N1IH47 S4P460 E3UPC9 A0A194PUA5 A0A2J7PK69 A0A0L7L5C2 A0A1Y1KRE6 A0A1W4X4U1 D2A131 A0A310SAE1 A0A0M9ACC3 A0A067R3S4 E2BWP0 E1ZZF1 A0A154PPB0 A0A026VYY9 A0A0L7QRY1 A0A087ZUZ4 A0A2A3E792 A0A232FG73 K7IR56 J3JVV9 A0A195ECF5 F4WNQ8 A0A195FVM0 A0A151IP55 Q7Q3Z0 A0A240PKQ6 A0A158NVP7 A0A1Y9HE26 A0A182GWG8 A0A084WGW4 A0A1L8DVQ4 A0A182Y069 A0A023EIU6 A0A240PMR3 A0A182WU42 A0A182L1H6 A0A182HUQ4 A0A2M3ZJV9 A0A0L0BUR8 A0A182UN56 A0A2M4BZ90 A0A1Y9H2Y4 A0A1Y9IW44 A0A1Y9G9G4 A0A2M4AQ60 A0A1Y9H9J4 A0A182UDH9 A0A2M4BZU3 W5JMH8 A0A182MIT9 A0A240PL21 A0A1I8N5Y8 U5EV61 A0A1B6D6Q7 A0A1Q3FDM4 A0A0K8TSD5 A0A1B6JFD7 B0X6Q2 A0A1B6FXS1 A0A1J1INN1 A0A1B6M0G6 A0A1I8QF50 A0A2S2Q9P7 B3N7D6 B4NXH0 B4HYQ3 B4Q786 E0VZF3 Q9VLI9 Q29JT8 B4N7T0 A0A0V0GAK5 A0A023EJV3 A0A1A9XEF1 A0A1B0AYT9 A0A3B0JL02 A0A1A9UYL4 A0A0P4VT81 T1HV27 A0A2S2PI94 G8JKE9 A0A069DQ86 B4JDZ6 B3MLY8 A0A2R7X2M3 B4LSE5 A0A2H8TPB8 A0A1W4W084 A0A0A1WTN3 A0A1B0DEK5 W8C8H1 A0A146KSS1
Pubmed
19121390
28756777
22118469
26354079
23622113
26227816
+ More
28004739 18362917 19820115 24845553 20798317 24508170 30249741 28648823 20075255 22516182 23537049 21719571 12364791 14747013 17210077 21347285 26483478 24438588 25244985 24945155 20966253 26108605 20920257 23761445 25315136 26369729 17994087 17550304 22936249 20566863 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 27129103 22679863 26334808 25830018 24495485 26823975
28004739 18362917 19820115 24845553 20798317 24508170 30249741 28648823 20075255 22516182 23537049 21719571 12364791 14747013 17210077 21347285 26483478 24438588 25244985 24945155 20966253 26108605 20920257 23761445 25315136 26369729 17994087 17550304 22936249 20566863 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 27129103 22679863 26334808 25830018 24495485 26823975
EMBL
BABH01038639
GDQN01009800
JAT81254.1
ODYU01011928
SOQ57954.1
KZ149931
+ More
PZC77340.1 NWSH01001319 PCG71714.1 AGBW02010072 OWR49237.1 KQ460642 KPJ13335.1 GAIX01006139 JAA86421.1 HM536613 ADO95152.1 KQ459592 KPI96902.1 NEVH01024940 PNF16731.1 JTDY01002923 KOB70489.1 GEZM01075758 GEZM01075757 GEZM01075756 JAV63972.1 KQ971338 EFA01590.1 KQ762207 OAD56031.1 KQ435701 KOX80373.1 KK852723 KDR17693.1 GL451165 EFN79895.1 GL435343 EFN73443.1 KQ435007 KZC13709.1 KK107558 QOIP01000012 EZA48987.1 RLU15976.1 KQ414766 KOC61383.1 KZ288345 PBC27633.1 NNAY01000242 OXU29766.1 APGK01018904 BT127377 KB740085 KB632303 AEE62339.1 ENN81539.1 ERL91805.1 KQ979074 KYN22791.1 GL888237 EGI64251.1 KQ981215 KYN44508.1 KQ976892 KYN07231.1 AAAB01008964 EAA12485.1 ADTU01002895 JXUM01093119 KQ564035 KXJ72894.1 ATLV01023763 KE525346 KFB49458.1 GFDF01003572 JAV10512.1 GAPW01004220 JAC09378.1 APCN01002471 GGFM01008060 MBW28811.1 JRES01001431 KNC22954.1 GGFJ01009258 MBW58399.1 GGFK01009578 MBW42899.1 AXCN02000765 GGFJ01009260 MBW58401.1 ADMH02000622 ETN65582.1 AXCM01005603 GANO01002022 JAB57849.1 GEDC01015930 JAS21368.1 GFDL01009392 JAV25653.1 GDAI01000336 JAI17267.1 GECU01009817 JAS97889.1 DS232421 EDS41535.1 GECZ01014779 JAS54990.1 CVRI01000054 CRL00702.1 GEBQ01010566 JAT29411.1 GGMS01005260 MBY74463.1 CH954177 EDV58287.1 CM000157 EDW88561.1 CH480818 EDW52183.1 CM000361 CM002910 EDX04280.1 KMY89134.1 DS235852 EEB18759.1 AE014134 BT120293 AAF52701.1 ADC41873.1 CH379062 EAL32873.1 CH964214 EDW80419.1 GECL01001231 JAP04893.1 GAPW01004236 JAC09362.1 JXJN01005947 OUUW01000006 SPP81493.1 GDKW01000781 JAI55814.1 ACPB03023272 ACPB03023273 GGMR01016540 MBY29159.1 JP593078 AER92463.1 GBGD01002854 JAC86035.1 CH916368 EDW03516.1 CH902620 EDV31816.1 KK856587 PTY26057.1 CH940649 EDW63753.1 GFXV01004191 MBW15996.1 GBXI01012442 JAD01850.1 AJVK01032881 GAMC01007466 JAB99089.1 GDHC01020382 JAP98246.1
PZC77340.1 NWSH01001319 PCG71714.1 AGBW02010072 OWR49237.1 KQ460642 KPJ13335.1 GAIX01006139 JAA86421.1 HM536613 ADO95152.1 KQ459592 KPI96902.1 NEVH01024940 PNF16731.1 JTDY01002923 KOB70489.1 GEZM01075758 GEZM01075757 GEZM01075756 JAV63972.1 KQ971338 EFA01590.1 KQ762207 OAD56031.1 KQ435701 KOX80373.1 KK852723 KDR17693.1 GL451165 EFN79895.1 GL435343 EFN73443.1 KQ435007 KZC13709.1 KK107558 QOIP01000012 EZA48987.1 RLU15976.1 KQ414766 KOC61383.1 KZ288345 PBC27633.1 NNAY01000242 OXU29766.1 APGK01018904 BT127377 KB740085 KB632303 AEE62339.1 ENN81539.1 ERL91805.1 KQ979074 KYN22791.1 GL888237 EGI64251.1 KQ981215 KYN44508.1 KQ976892 KYN07231.1 AAAB01008964 EAA12485.1 ADTU01002895 JXUM01093119 KQ564035 KXJ72894.1 ATLV01023763 KE525346 KFB49458.1 GFDF01003572 JAV10512.1 GAPW01004220 JAC09378.1 APCN01002471 GGFM01008060 MBW28811.1 JRES01001431 KNC22954.1 GGFJ01009258 MBW58399.1 GGFK01009578 MBW42899.1 AXCN02000765 GGFJ01009260 MBW58401.1 ADMH02000622 ETN65582.1 AXCM01005603 GANO01002022 JAB57849.1 GEDC01015930 JAS21368.1 GFDL01009392 JAV25653.1 GDAI01000336 JAI17267.1 GECU01009817 JAS97889.1 DS232421 EDS41535.1 GECZ01014779 JAS54990.1 CVRI01000054 CRL00702.1 GEBQ01010566 JAT29411.1 GGMS01005260 MBY74463.1 CH954177 EDV58287.1 CM000157 EDW88561.1 CH480818 EDW52183.1 CM000361 CM002910 EDX04280.1 KMY89134.1 DS235852 EEB18759.1 AE014134 BT120293 AAF52701.1 ADC41873.1 CH379062 EAL32873.1 CH964214 EDW80419.1 GECL01001231 JAP04893.1 GAPW01004236 JAC09362.1 JXJN01005947 OUUW01000006 SPP81493.1 GDKW01000781 JAI55814.1 ACPB03023272 ACPB03023273 GGMR01016540 MBY29159.1 JP593078 AER92463.1 GBGD01002854 JAC86035.1 CH916368 EDW03516.1 CH902620 EDV31816.1 KK856587 PTY26057.1 CH940649 EDW63753.1 GFXV01004191 MBW15996.1 GBXI01012442 JAD01850.1 AJVK01032881 GAMC01007466 JAB99089.1 GDHC01020382 JAP98246.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000235965
+ More
UP000037510 UP000192223 UP000007266 UP000053105 UP000027135 UP000008237 UP000000311 UP000076502 UP000053097 UP000279307 UP000053825 UP000005203 UP000242457 UP000215335 UP000002358 UP000019118 UP000030742 UP000078492 UP000007755 UP000078541 UP000078542 UP000007062 UP000075881 UP000005205 UP000075900 UP000069940 UP000249989 UP000030765 UP000076408 UP000075880 UP000076407 UP000075882 UP000075840 UP000037069 UP000075903 UP000075884 UP000075920 UP000069272 UP000075886 UP000075902 UP000000673 UP000075883 UP000075885 UP000095301 UP000002320 UP000183832 UP000095300 UP000008711 UP000002282 UP000001292 UP000000304 UP000009046 UP000000803 UP000001819 UP000007798 UP000092443 UP000092460 UP000268350 UP000078200 UP000015103 UP000001070 UP000007801 UP000008792 UP000192221 UP000092462
UP000037510 UP000192223 UP000007266 UP000053105 UP000027135 UP000008237 UP000000311 UP000076502 UP000053097 UP000279307 UP000053825 UP000005203 UP000242457 UP000215335 UP000002358 UP000019118 UP000030742 UP000078492 UP000007755 UP000078541 UP000078542 UP000007062 UP000075881 UP000005205 UP000075900 UP000069940 UP000249989 UP000030765 UP000076408 UP000075880 UP000076407 UP000075882 UP000075840 UP000037069 UP000075903 UP000075884 UP000075920 UP000069272 UP000075886 UP000075902 UP000000673 UP000075883 UP000075885 UP000095301 UP000002320 UP000183832 UP000095300 UP000008711 UP000002282 UP000001292 UP000000304 UP000009046 UP000000803 UP000001819 UP000007798 UP000092443 UP000092460 UP000268350 UP000078200 UP000015103 UP000001070 UP000007801 UP000008792 UP000192221 UP000092462
Pfam
PF04099 Sybindin
ProteinModelPortal
H9J8M2
A0A1E1W2S6
A0A2H1WZY7
A0A2W1BX02
A0A2A4JJE4
A0A212F677
+ More
A0A0N1IH47 S4P460 E3UPC9 A0A194PUA5 A0A2J7PK69 A0A0L7L5C2 A0A1Y1KRE6 A0A1W4X4U1 D2A131 A0A310SAE1 A0A0M9ACC3 A0A067R3S4 E2BWP0 E1ZZF1 A0A154PPB0 A0A026VYY9 A0A0L7QRY1 A0A087ZUZ4 A0A2A3E792 A0A232FG73 K7IR56 J3JVV9 A0A195ECF5 F4WNQ8 A0A195FVM0 A0A151IP55 Q7Q3Z0 A0A240PKQ6 A0A158NVP7 A0A1Y9HE26 A0A182GWG8 A0A084WGW4 A0A1L8DVQ4 A0A182Y069 A0A023EIU6 A0A240PMR3 A0A182WU42 A0A182L1H6 A0A182HUQ4 A0A2M3ZJV9 A0A0L0BUR8 A0A182UN56 A0A2M4BZ90 A0A1Y9H2Y4 A0A1Y9IW44 A0A1Y9G9G4 A0A2M4AQ60 A0A1Y9H9J4 A0A182UDH9 A0A2M4BZU3 W5JMH8 A0A182MIT9 A0A240PL21 A0A1I8N5Y8 U5EV61 A0A1B6D6Q7 A0A1Q3FDM4 A0A0K8TSD5 A0A1B6JFD7 B0X6Q2 A0A1B6FXS1 A0A1J1INN1 A0A1B6M0G6 A0A1I8QF50 A0A2S2Q9P7 B3N7D6 B4NXH0 B4HYQ3 B4Q786 E0VZF3 Q9VLI9 Q29JT8 B4N7T0 A0A0V0GAK5 A0A023EJV3 A0A1A9XEF1 A0A1B0AYT9 A0A3B0JL02 A0A1A9UYL4 A0A0P4VT81 T1HV27 A0A2S2PI94 G8JKE9 A0A069DQ86 B4JDZ6 B3MLY8 A0A2R7X2M3 B4LSE5 A0A2H8TPB8 A0A1W4W084 A0A0A1WTN3 A0A1B0DEK5 W8C8H1 A0A146KSS1
A0A0N1IH47 S4P460 E3UPC9 A0A194PUA5 A0A2J7PK69 A0A0L7L5C2 A0A1Y1KRE6 A0A1W4X4U1 D2A131 A0A310SAE1 A0A0M9ACC3 A0A067R3S4 E2BWP0 E1ZZF1 A0A154PPB0 A0A026VYY9 A0A0L7QRY1 A0A087ZUZ4 A0A2A3E792 A0A232FG73 K7IR56 J3JVV9 A0A195ECF5 F4WNQ8 A0A195FVM0 A0A151IP55 Q7Q3Z0 A0A240PKQ6 A0A158NVP7 A0A1Y9HE26 A0A182GWG8 A0A084WGW4 A0A1L8DVQ4 A0A182Y069 A0A023EIU6 A0A240PMR3 A0A182WU42 A0A182L1H6 A0A182HUQ4 A0A2M3ZJV9 A0A0L0BUR8 A0A182UN56 A0A2M4BZ90 A0A1Y9H2Y4 A0A1Y9IW44 A0A1Y9G9G4 A0A2M4AQ60 A0A1Y9H9J4 A0A182UDH9 A0A2M4BZU3 W5JMH8 A0A182MIT9 A0A240PL21 A0A1I8N5Y8 U5EV61 A0A1B6D6Q7 A0A1Q3FDM4 A0A0K8TSD5 A0A1B6JFD7 B0X6Q2 A0A1B6FXS1 A0A1J1INN1 A0A1B6M0G6 A0A1I8QF50 A0A2S2Q9P7 B3N7D6 B4NXH0 B4HYQ3 B4Q786 E0VZF3 Q9VLI9 Q29JT8 B4N7T0 A0A0V0GAK5 A0A023EJV3 A0A1A9XEF1 A0A1B0AYT9 A0A3B0JL02 A0A1A9UYL4 A0A0P4VT81 T1HV27 A0A2S2PI94 G8JKE9 A0A069DQ86 B4JDZ6 B3MLY8 A0A2R7X2M3 B4LSE5 A0A2H8TPB8 A0A1W4W084 A0A0A1WTN3 A0A1B0DEK5 W8C8H1 A0A146KSS1
PDB
2J3T
E-value=9.73862e-73,
Score=692
Ontologies
GO
PANTHER
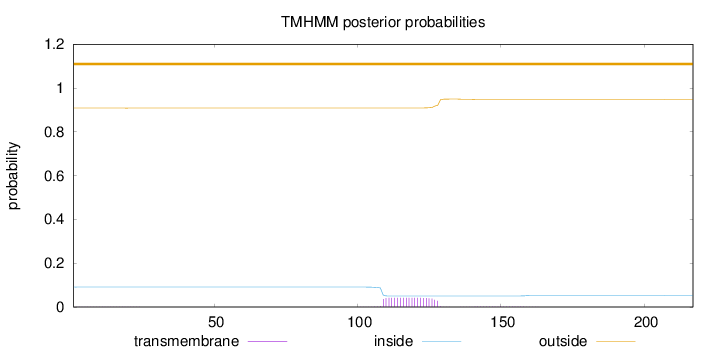
Topology
Length:
217
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.86285
Exp number, first 60 AAs:
0.01638
Total prob of N-in:
0.09158
outside
1 - 217
Population Genetic Test Statistics
Pi
17.028144
Theta
17.17568
Tajima's D
0.090352
CLR
0
CSRT
0.399830008499575
Interpretation
Uncertain
