Gene
KWMTBOMO07539
Pre Gene Modal
BGIBMGA005868
Annotation
PREDICTED:_sphingosine-1-phosphate_phosphatase_1-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.942
Sequence
CDS
ATGTGGGAGAATACGATAAAATACTTAAAAGATCCATTATTAGTTATTAAAGTACAAAATTTTTTCGGCGTCACTTATAAATCTAACAGACAGATCAGTGATACGAAGAGCGAACAGTCTAAAGTGATAAATTCAGAGAGACTAGACCGCCTGAGAGAAGAATGCGATATCAAACAGCACAAGAGGATACCAAGCAACATATCTAACAGCTCACAGTCCTCGTACGACACTGATACCTCTGGAGAAACAACGACGAAAGACGATCTCGGGCTCTCAATCAATAACAAATTCTGGTACTACTTGTTTGTACTCGGAACTGAGTTAGGAGATGAGATATTCTACGCTACATTCATACCGTTTTGGTTTTGGAATATTGACGGCGCCGTAGGACGGCGAGTCGTATTGGTTTGGACTGTCGTTATGTACATTGGACAAGGTTTTAAGGATATAATTCGATGGCCTCGCCCTGGTTACCCAGTAAAGAAGTTACAGCAGAAATGGGCTATAGAATACGGTATGCCCTCCACTCACGCCATGGTGGGAGTGTGCATGCCATTCTCAGTATTACTCTATACAATGGACAGGTATATTTACCCAGTCCACTGGGGGATACTGATAGCAGTGTCATGGTGTACCCTGATCTGTGTTAGTAGAGTATATCTGGGAATGCATAGTGTGTTGGATATTGCAGCAGGCTTGCTCCTCGCTTCATCTCTGCTATTCATCCTGATACCTGCGGTAGACCGACTGGACGGTTTCCTCCTCACGTGGGAGTGTGCTCCCGTGTTGGTCATCACACTCTCAATACTTGTCATCGTGTTTCATCCTAATGCCGATAAATGGACACCAACTAGAGGCGACACGACCATGATCGTGAGCGTGTGCGCGGGCATCCTGACCGGCTCCTGGACCAACTACCGGCTGGGGAACATGACGGCCGACGACCGGGGCCCCCCCTACCGCATCATATGGCCCTCCGTGGAGATGCTGGGCTGCACCATCCTCAGGACGACGCTGGGCTTCTGCGGCGTGTTGGCCACCAGGGCCATCGGCAAGTCAGTGTCGTACGCCTTCATCTGCGCGCTGCTAGGCAAAGACAAGAACGAATTGAGGAATTCTGAAGATAGTTTAGACAATAAGCACAAGGTGACAGTGGAGCTGTGCTACAAGTACTTCACGTACGCGATGGTGGGCTTCAACACCACGTTCGTGTTCCCCAACGTGTTCTCGCTGCTCAGCATCAACAGGCCCACCTACTACACAGAGGTCTAG
Protein
MWENTIKYLKDPLLVIKVQNFFGVTYKSNRQISDTKSEQSKVINSERLDRLREECDIKQHKRIPSNISNSSQSSYDTDTSGETTTKDDLGLSINNKFWYYLFVLGTELGDEIFYATFIPFWFWNIDGAVGRRVVLVWTVVMYIGQGFKDIIRWPRPGYPVKKLQQKWAIEYGMPSTHAMVGVCMPFSVLLYTMDRYIYPVHWGILIAVSWCTLICVSRVYLGMHSVLDIAAGLLLASSLLFILIPAVDRLDGFLLTWECAPVLVITLSILVIVFHPNADKWTPTRGDTTMIVSVCAGILTGSWTNYRLGNMTADDRGPPYRIIWPSVEMLGCTILRTTLGFCGVLATRAIGKSVSYAFICALLGKDKNELRNSEDSLDNKHKVTVELCYKYFTYAMVGFNTTFVFPNVFSLLSINRPTYYTEV
Summary
Uniprot
H9J8M6
A0A2W1BXF9
A0A2A4JD15
A0A1E1W4H8
A0A3S2TJ63
A0A0L7LI83
+ More
A0A0N1PHX7 A0A212F695 A0A194Q0E9 Q16S24 A0A182H4Q5 A0A023EUA2 A0A1B0DM25 A0A1L8DYG5 A0A1L8DYC7 A0A067R1I7 A0A182YFV5 A0A2M4BPB5 A0A182IJ10 A0A2M4CN55 A0A0P4VK60 A0A182F279 A0A182LUV1 A0A2J7R4Y5 A0A182Q369 A0A182R3X0 A0A2M4ARI3 A0A2M4ARH7 A0A023F0J4 A0A182NBH8 A0A084VH52 A0A182JYB0 A0A182VTY7 A0A182PDC2 A0A0V0G394 A0A232FH05 A0A182HI86 A0A182XN44 A0A182LM78 A0A182UC94 K7IZX9 A0A182UVH7 W5JAX1 A0A1Y1JV79 T1I6Q3 D6X060 A0A224XF97 A0A1B6D3K7 N6TDQ4 A0A069DU16 Q7Q1Q6 A0A2R7WC47 B0WQ16 A0A0A9WK25 A0A2A3EG62 V9IKN0 A0A087ZYX3 A0A0C9R951 E2C638 A0A0M9A1B4 A0A1B0CE11 A0A1B6LUV2 A0A310SUE4 A0A0L7QY32 A0A1B6HP62 A0A158NYF8 A0A1B6GRT7 E0VN03 A0A195F6H5 A0A026W7S2 A0A151JF26 A0A154PD58 T1ISK6 A0A0P4WHW6 E2AW89 A0A1B6GRA3 A0A226EP55 E9JD56 A0A164L7I3 A0A0P5LAD4 A0A0P6BNE5 E9H133 A0A0N8DZ58 A0A087U7W7 A0A0P6HE68 A0A2R5LG84 A0A1S3ILD0 A0A0P5EN76 A0A2S2NIJ6 J9K3Q3 A0A2H8TW14 A0A2S2PY91 R7TAT3 A0A293M1D1 A0A2H8TT34 A0A267EVE4
A0A0N1PHX7 A0A212F695 A0A194Q0E9 Q16S24 A0A182H4Q5 A0A023EUA2 A0A1B0DM25 A0A1L8DYG5 A0A1L8DYC7 A0A067R1I7 A0A182YFV5 A0A2M4BPB5 A0A182IJ10 A0A2M4CN55 A0A0P4VK60 A0A182F279 A0A182LUV1 A0A2J7R4Y5 A0A182Q369 A0A182R3X0 A0A2M4ARI3 A0A2M4ARH7 A0A023F0J4 A0A182NBH8 A0A084VH52 A0A182JYB0 A0A182VTY7 A0A182PDC2 A0A0V0G394 A0A232FH05 A0A182HI86 A0A182XN44 A0A182LM78 A0A182UC94 K7IZX9 A0A182UVH7 W5JAX1 A0A1Y1JV79 T1I6Q3 D6X060 A0A224XF97 A0A1B6D3K7 N6TDQ4 A0A069DU16 Q7Q1Q6 A0A2R7WC47 B0WQ16 A0A0A9WK25 A0A2A3EG62 V9IKN0 A0A087ZYX3 A0A0C9R951 E2C638 A0A0M9A1B4 A0A1B0CE11 A0A1B6LUV2 A0A310SUE4 A0A0L7QY32 A0A1B6HP62 A0A158NYF8 A0A1B6GRT7 E0VN03 A0A195F6H5 A0A026W7S2 A0A151JF26 A0A154PD58 T1ISK6 A0A0P4WHW6 E2AW89 A0A1B6GRA3 A0A226EP55 E9JD56 A0A164L7I3 A0A0P5LAD4 A0A0P6BNE5 E9H133 A0A0N8DZ58 A0A087U7W7 A0A0P6HE68 A0A2R5LG84 A0A1S3ILD0 A0A0P5EN76 A0A2S2NIJ6 J9K3Q3 A0A2H8TW14 A0A2S2PY91 R7TAT3 A0A293M1D1 A0A2H8TT34 A0A267EVE4
Pubmed
19121390
28756777
26227816
26354079
22118469
17510324
+ More
26483478 24945155 24845553 25244985 27129103 25474469 24438588 28648823 20966253 20075255 20920257 23761445 28004739 18362917 19820115 23537049 26334808 12364791 25401762 26823975 20798317 21347285 20566863 24508170 30249741 21282665 21292972 23254933
26483478 24945155 24845553 25244985 27129103 25474469 24438588 28648823 20966253 20075255 20920257 23761445 28004739 18362917 19820115 23537049 26334808 12364791 25401762 26823975 20798317 21347285 20566863 24508170 30249741 21282665 21292972 23254933
EMBL
BABH01038663
KZ149931
PZC77356.1
NWSH01002048
PCG69312.1
GDQN01009190
+ More
JAT81864.1 RSAL01000101 RVE47506.1 JTDY01000967 KOB75253.1 KQ460642 KPJ13327.1 AGBW02010072 OWR49229.1 KQ459592 KPI96890.1 CH477688 EAT37263.1 JXUM01003908 KQ560165 KXJ84052.1 GAPW01001594 JAC12004.1 AJVK01006872 GFDF01002625 JAV11459.1 GFDF01002624 JAV11460.1 KK852777 KDR16780.1 GGFJ01005785 MBW54926.1 GGFL01002566 MBW66744.1 GDKW01001169 JAI55426.1 AXCM01002025 NEVH01007399 PNF35891.1 AXCN02000616 GGFK01010085 MBW43406.1 GGFK01010084 MBW43405.1 GBBI01003742 JAC14970.1 ATLV01013143 KE524841 KFB37296.1 GECL01003653 JAP02471.1 NNAY01000216 OXU29965.1 APCN01005655 ADMH02001762 ETN61151.1 GEZM01100142 JAV52993.1 ACPB03010942 KQ971371 EFA10076.1 GFTR01005269 JAW11157.1 GEDC01017062 JAS20236.1 APGK01041789 KB740997 KB632100 ENN75878.1 ERL88819.1 GBGD01001628 JAC87261.1 AAAB01008980 EAA14540.3 KK854439 PTY15865.1 DS232032 EDS32660.1 GBHO01038399 GBHO01038398 GBRD01000663 GDHC01018400 JAG05205.1 JAG05206.1 JAG65158.1 JAQ00229.1 KZ288269 PBC29991.1 JR052380 AEY61688.1 GBYB01012906 GBYB01012907 GBYB01012908 GBYB01012909 JAG82673.1 JAG82674.1 JAG82675.1 JAG82676.1 GL452916 EFN76581.1 KQ435765 KOX75320.1 AJWK01008481 AJWK01008482 GEBQ01012530 JAT27447.1 KQ759870 OAD62429.1 KQ414697 KOC63517.1 GECU01031229 JAS76477.1 ADTU01003789 GECZ01004639 JAS65130.1 DS235331 EEB14759.1 KQ981768 KYN35986.1 KK107419 QOIP01000003 EZA51089.1 RLU24585.1 KQ979032 KYN24328.1 KQ434875 KZC09782.1 JH431434 GDRN01088209 GDRN01088208 JAI60895.1 GL443262 EFN62311.1 GECZ01004803 JAS64966.1 LNIX01000002 OXA59413.1 GL771866 EFZ09211.1 LRGB01003164 KZS03856.1 GDIQ01179678 JAK72047.1 GDIP01018924 JAM84791.1 GL732582 EFX74592.1 GDIQ01077604 JAN17133.1 KK118620 KFM73456.1 GDIQ01020531 JAN74206.1 GGLE01004353 MBY08479.1 GDIP01143958 JAJ79444.1 GGMR01004329 MBY16948.1 ABLF02029160 ABLF02029163 GFXV01006678 MBW18483.1 GGMS01000689 MBY69892.1 AMQN01015504 KB311970 ELT88124.1 GFWV01009397 MAA34126.1 GFXV01005294 MBW17099.1 NIVC01001645 PAA65500.1
JAT81864.1 RSAL01000101 RVE47506.1 JTDY01000967 KOB75253.1 KQ460642 KPJ13327.1 AGBW02010072 OWR49229.1 KQ459592 KPI96890.1 CH477688 EAT37263.1 JXUM01003908 KQ560165 KXJ84052.1 GAPW01001594 JAC12004.1 AJVK01006872 GFDF01002625 JAV11459.1 GFDF01002624 JAV11460.1 KK852777 KDR16780.1 GGFJ01005785 MBW54926.1 GGFL01002566 MBW66744.1 GDKW01001169 JAI55426.1 AXCM01002025 NEVH01007399 PNF35891.1 AXCN02000616 GGFK01010085 MBW43406.1 GGFK01010084 MBW43405.1 GBBI01003742 JAC14970.1 ATLV01013143 KE524841 KFB37296.1 GECL01003653 JAP02471.1 NNAY01000216 OXU29965.1 APCN01005655 ADMH02001762 ETN61151.1 GEZM01100142 JAV52993.1 ACPB03010942 KQ971371 EFA10076.1 GFTR01005269 JAW11157.1 GEDC01017062 JAS20236.1 APGK01041789 KB740997 KB632100 ENN75878.1 ERL88819.1 GBGD01001628 JAC87261.1 AAAB01008980 EAA14540.3 KK854439 PTY15865.1 DS232032 EDS32660.1 GBHO01038399 GBHO01038398 GBRD01000663 GDHC01018400 JAG05205.1 JAG05206.1 JAG65158.1 JAQ00229.1 KZ288269 PBC29991.1 JR052380 AEY61688.1 GBYB01012906 GBYB01012907 GBYB01012908 GBYB01012909 JAG82673.1 JAG82674.1 JAG82675.1 JAG82676.1 GL452916 EFN76581.1 KQ435765 KOX75320.1 AJWK01008481 AJWK01008482 GEBQ01012530 JAT27447.1 KQ759870 OAD62429.1 KQ414697 KOC63517.1 GECU01031229 JAS76477.1 ADTU01003789 GECZ01004639 JAS65130.1 DS235331 EEB14759.1 KQ981768 KYN35986.1 KK107419 QOIP01000003 EZA51089.1 RLU24585.1 KQ979032 KYN24328.1 KQ434875 KZC09782.1 JH431434 GDRN01088209 GDRN01088208 JAI60895.1 GL443262 EFN62311.1 GECZ01004803 JAS64966.1 LNIX01000002 OXA59413.1 GL771866 EFZ09211.1 LRGB01003164 KZS03856.1 GDIQ01179678 JAK72047.1 GDIP01018924 JAM84791.1 GL732582 EFX74592.1 GDIQ01077604 JAN17133.1 KK118620 KFM73456.1 GDIQ01020531 JAN74206.1 GGLE01004353 MBY08479.1 GDIP01143958 JAJ79444.1 GGMR01004329 MBY16948.1 ABLF02029160 ABLF02029163 GFXV01006678 MBW18483.1 GGMS01000689 MBY69892.1 AMQN01015504 KB311970 ELT88124.1 GFWV01009397 MAA34126.1 GFXV01005294 MBW17099.1 NIVC01001645 PAA65500.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000037510
UP000053240
UP000007151
+ More
UP000053268 UP000008820 UP000069940 UP000249989 UP000092462 UP000027135 UP000076408 UP000075880 UP000069272 UP000075883 UP000235965 UP000075886 UP000075900 UP000075884 UP000030765 UP000075881 UP000075920 UP000075885 UP000215335 UP000075840 UP000076407 UP000075882 UP000075902 UP000002358 UP000075903 UP000000673 UP000015103 UP000007266 UP000019118 UP000030742 UP000007062 UP000002320 UP000242457 UP000005203 UP000008237 UP000053105 UP000092461 UP000053825 UP000005205 UP000009046 UP000078541 UP000053097 UP000279307 UP000078492 UP000076502 UP000000311 UP000198287 UP000076858 UP000000305 UP000054359 UP000085678 UP000007819 UP000014760 UP000215902
UP000053268 UP000008820 UP000069940 UP000249989 UP000092462 UP000027135 UP000076408 UP000075880 UP000069272 UP000075883 UP000235965 UP000075886 UP000075900 UP000075884 UP000030765 UP000075881 UP000075920 UP000075885 UP000215335 UP000075840 UP000076407 UP000075882 UP000075902 UP000002358 UP000075903 UP000000673 UP000015103 UP000007266 UP000019118 UP000030742 UP000007062 UP000002320 UP000242457 UP000005203 UP000008237 UP000053105 UP000092461 UP000053825 UP000005205 UP000009046 UP000078541 UP000053097 UP000279307 UP000078492 UP000076502 UP000000311 UP000198287 UP000076858 UP000000305 UP000054359 UP000085678 UP000007819 UP000014760 UP000215902
PRIDE
Pfam
PF01569 PAP2
SUPFAM
SSF48317
SSF48317
ProteinModelPortal
H9J8M6
A0A2W1BXF9
A0A2A4JD15
A0A1E1W4H8
A0A3S2TJ63
A0A0L7LI83
+ More
A0A0N1PHX7 A0A212F695 A0A194Q0E9 Q16S24 A0A182H4Q5 A0A023EUA2 A0A1B0DM25 A0A1L8DYG5 A0A1L8DYC7 A0A067R1I7 A0A182YFV5 A0A2M4BPB5 A0A182IJ10 A0A2M4CN55 A0A0P4VK60 A0A182F279 A0A182LUV1 A0A2J7R4Y5 A0A182Q369 A0A182R3X0 A0A2M4ARI3 A0A2M4ARH7 A0A023F0J4 A0A182NBH8 A0A084VH52 A0A182JYB0 A0A182VTY7 A0A182PDC2 A0A0V0G394 A0A232FH05 A0A182HI86 A0A182XN44 A0A182LM78 A0A182UC94 K7IZX9 A0A182UVH7 W5JAX1 A0A1Y1JV79 T1I6Q3 D6X060 A0A224XF97 A0A1B6D3K7 N6TDQ4 A0A069DU16 Q7Q1Q6 A0A2R7WC47 B0WQ16 A0A0A9WK25 A0A2A3EG62 V9IKN0 A0A087ZYX3 A0A0C9R951 E2C638 A0A0M9A1B4 A0A1B0CE11 A0A1B6LUV2 A0A310SUE4 A0A0L7QY32 A0A1B6HP62 A0A158NYF8 A0A1B6GRT7 E0VN03 A0A195F6H5 A0A026W7S2 A0A151JF26 A0A154PD58 T1ISK6 A0A0P4WHW6 E2AW89 A0A1B6GRA3 A0A226EP55 E9JD56 A0A164L7I3 A0A0P5LAD4 A0A0P6BNE5 E9H133 A0A0N8DZ58 A0A087U7W7 A0A0P6HE68 A0A2R5LG84 A0A1S3ILD0 A0A0P5EN76 A0A2S2NIJ6 J9K3Q3 A0A2H8TW14 A0A2S2PY91 R7TAT3 A0A293M1D1 A0A2H8TT34 A0A267EVE4
A0A0N1PHX7 A0A212F695 A0A194Q0E9 Q16S24 A0A182H4Q5 A0A023EUA2 A0A1B0DM25 A0A1L8DYG5 A0A1L8DYC7 A0A067R1I7 A0A182YFV5 A0A2M4BPB5 A0A182IJ10 A0A2M4CN55 A0A0P4VK60 A0A182F279 A0A182LUV1 A0A2J7R4Y5 A0A182Q369 A0A182R3X0 A0A2M4ARI3 A0A2M4ARH7 A0A023F0J4 A0A182NBH8 A0A084VH52 A0A182JYB0 A0A182VTY7 A0A182PDC2 A0A0V0G394 A0A232FH05 A0A182HI86 A0A182XN44 A0A182LM78 A0A182UC94 K7IZX9 A0A182UVH7 W5JAX1 A0A1Y1JV79 T1I6Q3 D6X060 A0A224XF97 A0A1B6D3K7 N6TDQ4 A0A069DU16 Q7Q1Q6 A0A2R7WC47 B0WQ16 A0A0A9WK25 A0A2A3EG62 V9IKN0 A0A087ZYX3 A0A0C9R951 E2C638 A0A0M9A1B4 A0A1B0CE11 A0A1B6LUV2 A0A310SUE4 A0A0L7QY32 A0A1B6HP62 A0A158NYF8 A0A1B6GRT7 E0VN03 A0A195F6H5 A0A026W7S2 A0A151JF26 A0A154PD58 T1ISK6 A0A0P4WHW6 E2AW89 A0A1B6GRA3 A0A226EP55 E9JD56 A0A164L7I3 A0A0P5LAD4 A0A0P6BNE5 E9H133 A0A0N8DZ58 A0A087U7W7 A0A0P6HE68 A0A2R5LG84 A0A1S3ILD0 A0A0P5EN76 A0A2S2NIJ6 J9K3Q3 A0A2H8TW14 A0A2S2PY91 R7TAT3 A0A293M1D1 A0A2H8TT34 A0A267EVE4
Ontologies
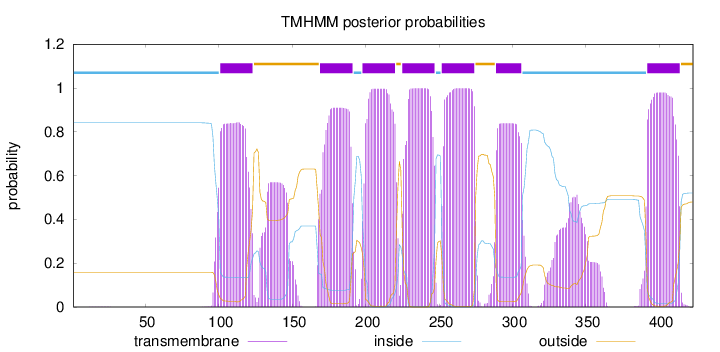
Topology
Length:
423
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
167.0894
Exp number, first 60 AAs:
0.00367
Total prob of N-in:
0.84150
inside
1 - 100
TMhelix
101 - 123
outside
124 - 168
TMhelix
169 - 191
inside
192 - 197
TMhelix
198 - 220
outside
221 - 224
TMhelix
225 - 247
inside
248 - 251
TMhelix
252 - 274
outside
275 - 288
TMhelix
289 - 306
inside
307 - 391
TMhelix
392 - 414
outside
415 - 423
Population Genetic Test Statistics
Pi
22.227502
Theta
17.864144
Tajima's D
0.17518
CLR
1.73018
CSRT
0.431878406079696
Interpretation
Uncertain
