Gene
KWMTBOMO07537
Pre Gene Modal
BGIBMGA005869
Annotation
PREDICTED:_DNA_replication_complex_GINS_protein_PSF1-like_[Plutella_xylostella]
Full name
DNA replication complex GINS protein PSF1
Alternative Name
GINS complex subunit 1
Location in the cell
Nuclear Reliability : 3.433
Sequence
CDS
ATGTTCGCAGAAAAAGTTATAGAACTAATTAAAGAAGTAGATAGAAATCCTGACACAATAGATCAATATAATGATGAAAAAATACGCCAAATAATAGAAGAAATGCATTTATTATTTAAATTGAACATGAATGATGCTAATGCTACAACAGAAAACAGATCATTGTGGACCACAGTTCAAGTAAGACATTCTGCCCTTGAACGCAATAAAAGGTGTATATTAGCCTATCTCTACAACAGAATGAAAAAAATTAAGACATTACGTTGGGAATTTGGTGCAGTACTACCTCCAGATATCAGAGAATTGCTTTCTGATGATGAATATCAATGGTTTTCAAAATACTCTGGAAATCTTGCTTTGTATATGAGATCTTTAGGCCAAGATATAGGATACAGTGGCATAGACTTGACTGAAAATTTAAAGCCCCCTAAATCATTGTTCATCGAAGTGCTTTGTGTTGTGGATTATGGTAAATTAGAACTTGAAGATGGTGATGTGGTTATGCTTAAGAAGAATAGCAGACATTATCTTCCTACTTCGGAATGTCAGACTTTAATACGACAGGGTATTCTGAAACAAATAATGTGA
Protein
MFAEKVIELIKEVDRNPDTIDQYNDEKIRQIIEEMHLLFKLNMNDANATTENRSLWTTVQVRHSALERNKRCILAYLYNRMKKIKTLRWEFGAVLPPDIRELLSDDEYQWFSKYSGNLALYMRSLGQDIGYSGIDLTENLKPPKSLFIEVLCVVDYGKLELEDGDVVMLKKNSRHYLPTSECQTLIRQGILKQIM
Summary
Description
Required for correct functioning of the GINS complex, a complex that plays an essential role in the initiation of DNA replication, and progression of DNA replication forks. GINS complex seems to bind preferentially to single-stranded DNA.
Subunit
Component of the GINS complex which is a heterotetramer of GINS1, GINS2, GINS3 and GINS4. Forms a stable subcomplex with GINS4. GINS complex interacts with DNA primase in vitro.
Similarity
Belongs to the GINS1/PSF1 family.
Keywords
Complete proteome
DNA replication
Nucleus
Reference proteome
Feature
chain DNA replication complex GINS protein PSF1
Uniprot
A0A1E1VYQ2
S4PFR2
A0A194PU91
A0A2W1BQR7
A0A0N1I944
A0A2A4JCZ6
+ More
A0A2H1VK42 A0A3S2NH85 A0A212F680 A0A0L7KXK1 C3XR78 A0A1S3IV78 A7SVK7 K1R2F6 A0A1I8PL06 W8C2C7 A0A067R829 A0A1I8NFK5 B3M8X8 H2ZEV3 A0A0L0BRK7 A0A0M4EY07 B4MGQ9 A0A3R7PNL8 A0A034WQ23 A0A0K8TW82 A0A3B0JXX8 A0A2B4SGN1 B3NB77 B4HVQ2 B4PD56 B4MN91 B4QLY5 A0A0J9RLF3 Q9W0I7 H2XZ76 B5DQY2 B4H5E4 A0A3M6TUG0 B4KXY1 A0A2P2I8F2 A0A369SLU1 B4IYK4 T2MD54 A0A0P6JH59 E9GIC5 A0A1W4VVE4 A0A0L8GFI9 D7EIS4 F7EGJ1 A0A3S0ZL89 R7VH77 A0A154PBC0 E0VNU1 Q7ZT47 G3WYP1 G1KPP9 A0A3B1J2V7 A0A2I4CGG4 A0A2A3EDQ7 A0A087ZYR6 A0A2T7NXD7 A0A1B0A065 M3XLH2 A0A0C9RDB2 A0A1A9VRC7 A0A1B0FAE8 A0A3Q0EF59 A0A151M1H2 A0A3Q2ZIN7 A0A3P9A9S4 W5NJA9 A0A3Q3DZB2 A0A1A9XDT7 A0A1B0B8G5 G3PER8 A0A060W7E6 B9ENG0 B5DES7 S4RZP9 A0A3N0XWV6 A0A3B3ZYG5 V9L7Z5 C1BJP7 A0A2U9CDS9 Q5XJQ9 A0A3B4EPW3 A0A1A9WCI0 A0A3B4WJ87 A0A3B4TF60 A0A3P8NKH4 A0A3P9B601 A0A0P6JEF3 A0A3B4GRK1 A0A3Q2VHN7 Q9CZ15 A0A0L7RDJ7 I3JJ64 A0A2Y9DKX3
A0A2H1VK42 A0A3S2NH85 A0A212F680 A0A0L7KXK1 C3XR78 A0A1S3IV78 A7SVK7 K1R2F6 A0A1I8PL06 W8C2C7 A0A067R829 A0A1I8NFK5 B3M8X8 H2ZEV3 A0A0L0BRK7 A0A0M4EY07 B4MGQ9 A0A3R7PNL8 A0A034WQ23 A0A0K8TW82 A0A3B0JXX8 A0A2B4SGN1 B3NB77 B4HVQ2 B4PD56 B4MN91 B4QLY5 A0A0J9RLF3 Q9W0I7 H2XZ76 B5DQY2 B4H5E4 A0A3M6TUG0 B4KXY1 A0A2P2I8F2 A0A369SLU1 B4IYK4 T2MD54 A0A0P6JH59 E9GIC5 A0A1W4VVE4 A0A0L8GFI9 D7EIS4 F7EGJ1 A0A3S0ZL89 R7VH77 A0A154PBC0 E0VNU1 Q7ZT47 G3WYP1 G1KPP9 A0A3B1J2V7 A0A2I4CGG4 A0A2A3EDQ7 A0A087ZYR6 A0A2T7NXD7 A0A1B0A065 M3XLH2 A0A0C9RDB2 A0A1A9VRC7 A0A1B0FAE8 A0A3Q0EF59 A0A151M1H2 A0A3Q2ZIN7 A0A3P9A9S4 W5NJA9 A0A3Q3DZB2 A0A1A9XDT7 A0A1B0B8G5 G3PER8 A0A060W7E6 B9ENG0 B5DES7 S4RZP9 A0A3N0XWV6 A0A3B3ZYG5 V9L7Z5 C1BJP7 A0A2U9CDS9 Q5XJQ9 A0A3B4EPW3 A0A1A9WCI0 A0A3B4WJ87 A0A3B4TF60 A0A3P8NKH4 A0A3P9B601 A0A0P6JEF3 A0A3B4GRK1 A0A3Q2VHN7 Q9CZ15 A0A0L7RDJ7 I3JJ64 A0A2Y9DKX3
Pubmed
23622113
26354079
28756777
22118469
26227816
18563158
+ More
17615350 22992520 24495485 24845553 25315136 17994087 26108605 25348373 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12481130 15114417 15632085 30382153 30042472 24065732 21292972 18362917 19820115 17495919 23254933 20566863 12730133 27762356 21709235 25329095 9215903 22293439 25069045 24755649 20433749 20431018 25463417 24402279 25186727 16141072 21183079
17615350 22992520 24495485 24845553 25315136 17994087 26108605 25348373 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12481130 15114417 15632085 30382153 30042472 24065732 21292972 18362917 19820115 17495919 23254933 20566863 12730133 27762356 21709235 25329095 9215903 22293439 25069045 24755649 20433749 20431018 25463417 24402279 25186727 16141072 21183079
EMBL
GDQN01011240
JAT79814.1
GAIX01003912
JAA88648.1
KQ459592
KPI96887.1
+ More
KZ149931 PZC77358.1 KQ460642 KPJ13324.1 NWSH01002048 PCG69310.1 ODYU01002744 SOQ40624.1 RSAL01000101 RVE47504.1 AGBW02010072 OWR49224.1 JTDY01004576 KOB67993.1 GG666456 EEN69196.1 DS469838 EDO32255.1 JH818849 EKC37719.1 GAMC01000538 JAC06018.1 KK852637 KDR19699.1 CH902618 EDV38922.1 JRES01001467 KNC22672.1 CP012525 ALC42970.1 CH940682 EDW57125.1 QCYY01002184 ROT72272.1 GAKP01002662 JAC56290.1 GDHF01033761 GDHF01019104 JAI18553.1 JAI33210.1 OUUW01000012 SPP86927.1 LSMT01000087 PFX28249.1 CH954178 EDV50130.1 CH480817 EDW50017.1 CM000159 EDW92804.2 CH963847 EDW73647.1 CM000363 EDX08771.1 CM002912 KMY96773.1 AE014296 BT025002 AAF47459.2 ABE01232.1 EAAA01002037 CH379070 EDY73969.1 CH479211 EDW32996.1 RCHS01002927 RMX44928.1 CH933809 EDW17653.1 IACF01004607 LAB70196.1 NOWV01000003 RDD47454.1 CH916366 EDV95514.1 HAAD01003817 CDG70049.1 GDIP01065810 GDIQ01007889 JAM37905.1 JAN86848.1 GL732546 EFX80795.1 KQ422015 KOF75776.1 DS497685 EFA12251.1 RQTK01000397 RUS80353.1 AMQN01003927 AMQN01026028 KB305852 KB292234 ELU00601.1 ELU17932.1 KQ434864 KZC09107.1 DS235354 EEB15047.1 BC072857 AB097168 CM004475 AAH72857.1 BAC66458.1 OCT77784.1 AEFK01029041 AEFK01029042 KZ288269 PBC29913.1 PZQS01000008 PVD25822.1 AFYH01143668 AFYH01143669 AFYH01143670 GBYB01004936 JAG74703.1 CCAG010008635 AKHW03006817 KYO18367.1 AHAT01001756 JXJN01009934 FR904369 CDQ61159.1 BT057185 ACM09057.1 AAMC01070527 BC168788 BC170763 BC170767 AAI68788.1 AAI70763.1 RJVU01057857 ROK15749.1 JW875782 AFP08299.1 BT074826 ACO09250.1 CP026257 AWP14741.1 BC083239 AAH83239.1 GEBF01004656 JAN98976.1 AK013116 KQ414613 KOC69062.1 AERX01045888
KZ149931 PZC77358.1 KQ460642 KPJ13324.1 NWSH01002048 PCG69310.1 ODYU01002744 SOQ40624.1 RSAL01000101 RVE47504.1 AGBW02010072 OWR49224.1 JTDY01004576 KOB67993.1 GG666456 EEN69196.1 DS469838 EDO32255.1 JH818849 EKC37719.1 GAMC01000538 JAC06018.1 KK852637 KDR19699.1 CH902618 EDV38922.1 JRES01001467 KNC22672.1 CP012525 ALC42970.1 CH940682 EDW57125.1 QCYY01002184 ROT72272.1 GAKP01002662 JAC56290.1 GDHF01033761 GDHF01019104 JAI18553.1 JAI33210.1 OUUW01000012 SPP86927.1 LSMT01000087 PFX28249.1 CH954178 EDV50130.1 CH480817 EDW50017.1 CM000159 EDW92804.2 CH963847 EDW73647.1 CM000363 EDX08771.1 CM002912 KMY96773.1 AE014296 BT025002 AAF47459.2 ABE01232.1 EAAA01002037 CH379070 EDY73969.1 CH479211 EDW32996.1 RCHS01002927 RMX44928.1 CH933809 EDW17653.1 IACF01004607 LAB70196.1 NOWV01000003 RDD47454.1 CH916366 EDV95514.1 HAAD01003817 CDG70049.1 GDIP01065810 GDIQ01007889 JAM37905.1 JAN86848.1 GL732546 EFX80795.1 KQ422015 KOF75776.1 DS497685 EFA12251.1 RQTK01000397 RUS80353.1 AMQN01003927 AMQN01026028 KB305852 KB292234 ELU00601.1 ELU17932.1 KQ434864 KZC09107.1 DS235354 EEB15047.1 BC072857 AB097168 CM004475 AAH72857.1 BAC66458.1 OCT77784.1 AEFK01029041 AEFK01029042 KZ288269 PBC29913.1 PZQS01000008 PVD25822.1 AFYH01143668 AFYH01143669 AFYH01143670 GBYB01004936 JAG74703.1 CCAG010008635 AKHW03006817 KYO18367.1 AHAT01001756 JXJN01009934 FR904369 CDQ61159.1 BT057185 ACM09057.1 AAMC01070527 BC168788 BC170763 BC170767 AAI68788.1 AAI70763.1 RJVU01057857 ROK15749.1 JW875782 AFP08299.1 BT074826 ACO09250.1 CP026257 AWP14741.1 BC083239 AAH83239.1 GEBF01004656 JAN98976.1 AK013116 KQ414613 KOC69062.1 AERX01045888
Proteomes
UP000053268
UP000053240
UP000218220
UP000283053
UP000007151
UP000037510
+ More
UP000001554 UP000085678 UP000001593 UP000005408 UP000095300 UP000027135 UP000095301 UP000007801 UP000007875 UP000037069 UP000092553 UP000008792 UP000283509 UP000268350 UP000225706 UP000008711 UP000001292 UP000002282 UP000007798 UP000000304 UP000000803 UP000008144 UP000001819 UP000008744 UP000275408 UP000009192 UP000253843 UP000001070 UP000000305 UP000192221 UP000053454 UP000007266 UP000002280 UP000271974 UP000014760 UP000076502 UP000009046 UP000186698 UP000007648 UP000001646 UP000018467 UP000192220 UP000242457 UP000005203 UP000245119 UP000092445 UP000008672 UP000078200 UP000092444 UP000189704 UP000050525 UP000264800 UP000265140 UP000018468 UP000261660 UP000092443 UP000092460 UP000007635 UP000193380 UP000087266 UP000008143 UP000245300 UP000261520 UP000246464 UP000261440 UP000091820 UP000261360 UP000261420 UP000265100 UP000265160 UP000261460 UP000264840 UP000000589 UP000053825 UP000005207 UP000248480
UP000001554 UP000085678 UP000001593 UP000005408 UP000095300 UP000027135 UP000095301 UP000007801 UP000007875 UP000037069 UP000092553 UP000008792 UP000283509 UP000268350 UP000225706 UP000008711 UP000001292 UP000002282 UP000007798 UP000000304 UP000000803 UP000008144 UP000001819 UP000008744 UP000275408 UP000009192 UP000253843 UP000001070 UP000000305 UP000192221 UP000053454 UP000007266 UP000002280 UP000271974 UP000014760 UP000076502 UP000009046 UP000186698 UP000007648 UP000001646 UP000018467 UP000192220 UP000242457 UP000005203 UP000245119 UP000092445 UP000008672 UP000078200 UP000092444 UP000189704 UP000050525 UP000264800 UP000265140 UP000018468 UP000261660 UP000092443 UP000092460 UP000007635 UP000193380 UP000087266 UP000008143 UP000245300 UP000261520 UP000246464 UP000261440 UP000091820 UP000261360 UP000261420 UP000265100 UP000265160 UP000261460 UP000264840 UP000000589 UP000053825 UP000005207 UP000248480
Pfam
PF05916 Sld5
SUPFAM
SSF158573
SSF158573
ProteinModelPortal
A0A1E1VYQ2
S4PFR2
A0A194PU91
A0A2W1BQR7
A0A0N1I944
A0A2A4JCZ6
+ More
A0A2H1VK42 A0A3S2NH85 A0A212F680 A0A0L7KXK1 C3XR78 A0A1S3IV78 A7SVK7 K1R2F6 A0A1I8PL06 W8C2C7 A0A067R829 A0A1I8NFK5 B3M8X8 H2ZEV3 A0A0L0BRK7 A0A0M4EY07 B4MGQ9 A0A3R7PNL8 A0A034WQ23 A0A0K8TW82 A0A3B0JXX8 A0A2B4SGN1 B3NB77 B4HVQ2 B4PD56 B4MN91 B4QLY5 A0A0J9RLF3 Q9W0I7 H2XZ76 B5DQY2 B4H5E4 A0A3M6TUG0 B4KXY1 A0A2P2I8F2 A0A369SLU1 B4IYK4 T2MD54 A0A0P6JH59 E9GIC5 A0A1W4VVE4 A0A0L8GFI9 D7EIS4 F7EGJ1 A0A3S0ZL89 R7VH77 A0A154PBC0 E0VNU1 Q7ZT47 G3WYP1 G1KPP9 A0A3B1J2V7 A0A2I4CGG4 A0A2A3EDQ7 A0A087ZYR6 A0A2T7NXD7 A0A1B0A065 M3XLH2 A0A0C9RDB2 A0A1A9VRC7 A0A1B0FAE8 A0A3Q0EF59 A0A151M1H2 A0A3Q2ZIN7 A0A3P9A9S4 W5NJA9 A0A3Q3DZB2 A0A1A9XDT7 A0A1B0B8G5 G3PER8 A0A060W7E6 B9ENG0 B5DES7 S4RZP9 A0A3N0XWV6 A0A3B3ZYG5 V9L7Z5 C1BJP7 A0A2U9CDS9 Q5XJQ9 A0A3B4EPW3 A0A1A9WCI0 A0A3B4WJ87 A0A3B4TF60 A0A3P8NKH4 A0A3P9B601 A0A0P6JEF3 A0A3B4GRK1 A0A3Q2VHN7 Q9CZ15 A0A0L7RDJ7 I3JJ64 A0A2Y9DKX3
A0A2H1VK42 A0A3S2NH85 A0A212F680 A0A0L7KXK1 C3XR78 A0A1S3IV78 A7SVK7 K1R2F6 A0A1I8PL06 W8C2C7 A0A067R829 A0A1I8NFK5 B3M8X8 H2ZEV3 A0A0L0BRK7 A0A0M4EY07 B4MGQ9 A0A3R7PNL8 A0A034WQ23 A0A0K8TW82 A0A3B0JXX8 A0A2B4SGN1 B3NB77 B4HVQ2 B4PD56 B4MN91 B4QLY5 A0A0J9RLF3 Q9W0I7 H2XZ76 B5DQY2 B4H5E4 A0A3M6TUG0 B4KXY1 A0A2P2I8F2 A0A369SLU1 B4IYK4 T2MD54 A0A0P6JH59 E9GIC5 A0A1W4VVE4 A0A0L8GFI9 D7EIS4 F7EGJ1 A0A3S0ZL89 R7VH77 A0A154PBC0 E0VNU1 Q7ZT47 G3WYP1 G1KPP9 A0A3B1J2V7 A0A2I4CGG4 A0A2A3EDQ7 A0A087ZYR6 A0A2T7NXD7 A0A1B0A065 M3XLH2 A0A0C9RDB2 A0A1A9VRC7 A0A1B0FAE8 A0A3Q0EF59 A0A151M1H2 A0A3Q2ZIN7 A0A3P9A9S4 W5NJA9 A0A3Q3DZB2 A0A1A9XDT7 A0A1B0B8G5 G3PER8 A0A060W7E6 B9ENG0 B5DES7 S4RZP9 A0A3N0XWV6 A0A3B3ZYG5 V9L7Z5 C1BJP7 A0A2U9CDS9 Q5XJQ9 A0A3B4EPW3 A0A1A9WCI0 A0A3B4WJ87 A0A3B4TF60 A0A3P8NKH4 A0A3P9B601 A0A0P6JEF3 A0A3B4GRK1 A0A3Q2VHN7 Q9CZ15 A0A0L7RDJ7 I3JJ64 A0A2Y9DKX3
PDB
2Q9Q
E-value=1.15009e-50,
Score=501
Ontologies
KEGG
GO
PANTHER
Topology
Subcellular location
Nucleus
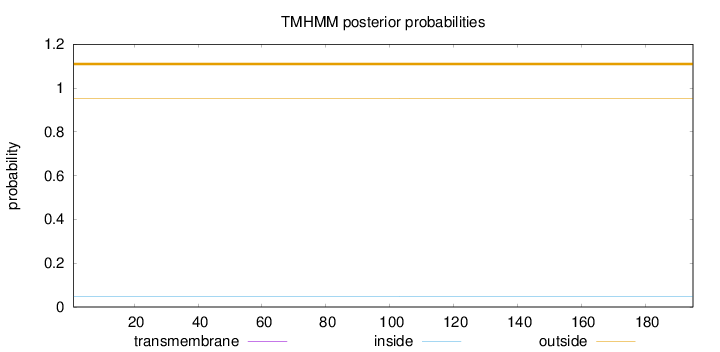
Length:
195
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04638
outside
1 - 195
Population Genetic Test Statistics
Pi
12.221252
Theta
17.755131
Tajima's D
-1.127817
CLR
0
CSRT
0.112944352782361
Interpretation
Uncertain
