Gene
KWMTBOMO07532
Pre Gene Modal
BGIBMGA005094
Annotation
PREDICTED:_ATP-binding_cassette_sub-family_G_member_1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.253
Sequence
CDS
ATGATCATGGGCATCAGGACACATGATGGTAATTGCGCGATGACGCCCGGCCTGAAAATGGAATCGAAAGGCACCCAGATACACTTACCGCTGATGGGGCGTGGGAGCAGAGGCGAGCTCAATGATAAAGTCACGTTCTCCCAGGGCAAAACCTTCACGGAGCAGCTCAATAATGGCCAAGTACTTGGTGGCTCCCAAATGAACCTATGCAATGGCGCTGCGGGTGCAGCAGGAATCACAACTCTCGTTCCCAAGATCCCAAATGGCTATGGAGATCAGAAAAAACCAATGGCGACTCTCACACACCTGCCAAAAAGACCTCCAGTGGACATAGAGTTCTCTGATTTATCCTATTCAGTCAGCGAGGGCAGGAAGAGAGGTTACAAGTCCTTGCTTAAATGTATCAATGGTACCTTTCGATCAGGGGAGCTGACAGCCATTATGGGACCTTCGGGGGCAGGGAAGAGCACACTCATGAACATTCTAGCCGGATATAAAACATCAAACGTCAGCGGATCAATCCTGATAAACGGAAAAGAAAGAAATTTGAGAAGATTCAGAAAACTATCCTGTTATATAATGCAGGATGACTGCCTTCTACCACATTTGACGGTCCGGGAGGCGATGTATGTATCAGCTAACCTGAAACTAGGGAAGGACATGACAGCTACAACAAAGAAGATAGTCATCGATGAAATATTAGAAATGCTCGGTCTTATCGAGGCCGGTGATACTAGGACCATAAATCTATCAGGAGGACAGAAAAAAAGACTATCAATAGCCTTGGAGCTAGTTAACAATCCACCGGTAATGTTCTTTGATGAACCAACTTCTGGATTGGATAGTTCCTCGTGTTTCCAATGCATATCATTATTAAAATCTTTGGCTAGAGGCGGCAGGACGATAATCTGTACTATCCATCAACCGTCTGCCCGATTATTTGAGATGTTTGACTTTTTATACACGTTGACCGATGGGCAGTGTATCTACCAAGGGAAGGTCACGGGTCTCGTGCCATTTTTGTCGTCGATGGGGCTCCACTGCCCAAGCTACCATAACCCTGCTGATTATTTAATGGAGGTAGCAAGCGGTGAACACGGTGAGTGGGTTCACAAGCTAGTAAGAGCCGTGAATAAAGGAAACTGCGGACGGGGACAGTCTTCTTCCACATCATCATCATCATCGTCCGTGCCTCAGAATGTAAATTACAATAATCAACCGAATAAAAATAATACTCAGAAAGAAGCTCTTTCTATAGTTGTTGACAAACCAACGTGTACTGTGATAGACATGTCAGAACCGGTACTCAATACCGAAAAGCAGAACATTCCAAACTCTACGAACCTCGCTCCTGTAACCTGCACCACTTCACTATTAGACTCCAGCGAGAGTTTCACAAAGAAGCCAGTTAAAACCGGTTTCCCGACGTCAGGGTGGAAGCAATTTTGGATCTTATTGAAGCGAACCTTCCGAAGTATTCTTAGAGACCAGACATTGACGCATTTAAGACTATGTTCGCATACGATCGTGGGATTGTTGATTGGCTTCCTCTATTACGATATAGGGAACGATGCCGCCAAAGTGATGAGCAATGCTGGATGCATTTTTTTTACCGTGATGTTCACCATGTTTACTGCCATGATGCCTACCATACTCACATTCCCGACAGAGATGAGCGTATTCGTGAGAGAACATTTAAATTACTGGTATTCGTTGAAGGCTTTCTATTTCGCAAAAACAATGGCAGATCTACCATTTCAGATAATATTCAGTGGCGTCTACGTGATCATAGTGTACTTCATGACGGGCCAGCCGATGCAGACCGATCGAGTGCTGATGTTCACGACCATCAACATACTCACGGCGCTAGTCTCCCAGTCTCTAGGACTTCTCATCGGTGCCGCCATGAAGATCGAGACTGGGGTGTACCTGGGACCGGTCACGACAATACCCGTCGTCTTATTCTCCGGTTTCTTCGTGAACTTCAACGCAATACCAAACTACCTTCGATGGCTGACGTACTTAAGCTATGTTAGATATGGCTTCGAAGGAGCCATGCTGTCGGTTTACGGGTTCGGAAGGGAGAAGCTGTATTGTTCGGAAGCATACTGCCATTTCCGGAACCCAGATCTCTTCTTGAAGGAAATGACGATGGATAAATCGAACTTTTGGATCGACGTTGGAGCTTTAAGCGCTCTGTTCGTCTTCATAAGAGTAATCTCATATTTAGTTCTAAGGTTTAAGTTGAAGATGATGCAATAA
Protein
MIMGIRTHDGNCAMTPGLKMESKGTQIHLPLMGRGSRGELNDKVTFSQGKTFTEQLNNGQVLGGSQMNLCNGAAGAAGITTLVPKIPNGYGDQKKPMATLTHLPKRPPVDIEFSDLSYSVSEGRKRGYKSLLKCINGTFRSGELTAIMGPSGAGKSTLMNILAGYKTSNVSGSILINGKERNLRRFRKLSCYIMQDDCLLPHLTVREAMYVSANLKLGKDMTATTKKIVIDEILEMLGLIEAGDTRTINLSGGQKKRLSIALELVNNPPVMFFDEPTSGLDSSSCFQCISLLKSLARGGRTIICTIHQPSARLFEMFDFLYTLTDGQCIYQGKVTGLVPFLSSMGLHCPSYHNPADYLMEVASGEHGEWVHKLVRAVNKGNCGRGQSSSTSSSSSSVPQNVNYNNQPNKNNTQKEALSIVVDKPTCTVIDMSEPVLNTEKQNIPNSTNLAPVTCTTSLLDSSESFTKKPVKTGFPTSGWKQFWILLKRTFRSILRDQTLTHLRLCSHTIVGLLIGFLYYDIGNDAAKVMSNAGCIFFTVMFTMFTAMMPTILTFPTEMSVFVREHLNYWYSLKAFYFAKTMADLPFQIIFSGVYVIIVYFMTGQPMQTDRVLMFTTINILTALVSQSLGLLIGAAMKIETGVYLGPVTTIPVVLFSGFFVNFNAIPNYLRWLTYLSYVRYGFEGAMLSVYGFGREKLYCSEAYCHFRNPDLFLKEMTMDKSNFWIDVGALSALFVFIRVISYLVLRFKLKMMQ
Summary
Uniprot
H9J6F2
A0A2H1WPU0
A0A212EU03
A0A194PVU7
A0A0N0PC93
E2A627
+ More
A0A158NYX8 A0A3L8DX23 V9ILI0 A0A1L8E4M1 A0A2A3EEV9 E2C0H1 F4W5H8 A0A088AN61 A0A026WAJ4 A0A151J3S1 A0A0N0U4H6 A0A0L7RDS6 K7IQL0 A0A0C9RP34 A0A067QKG8 A0A195BUM2 A0A0C9S320 A0A195D0X8 Q9VQY4 A0A2J7PQ37 A0A0Q5W964 B3N3Q6 A2RVF0 Q8MS50 A0A0J9QVL0 B4QA58 A0A154PC74 V5I802 D2A0Y3 Q9UAF0 A0A1J1IP34 A0A0R1DNL5 A0A1W4XW86 B4NYI9 B4I376 B3MLP2 A0A1W4XV81 A0A0P8XDZ5 A0A0T6BD08 A0A336LSN6 A0A336LS63 B4MDD2 A0A0Q9WLU5 B4N0N8 A0A0M4E9X6 A0A0U9HSJ3 A0A1Y1LFF6 A0A146LKB6 A0A146LYU6 A0A224X8K1 A0A023F379 A0A1B0CSL8 A0A0A9W0J1 A0A0P5U3B1 A0A0P6DCB0 A0A0P5I7T7 A0A0P5Q8Q9 A0A0P6G3P6 A0A0P6GAF5 A0A0P4YV08 E9HL04 A0A0P6F6A8 A0A0P5VQP2 A0A0A7ARN0 A0A0P5QQ66 A0A0P6AWT6 T1ITI8 A0A067QT28 A0A0P5X8I6 A0A232F3L7 A0A1S3IHP1 A0A2J7PQ84 K7J0G4 A0A2C9JEL2 A0A2C9JEL6 A0A2C9JEL7 A0A195D0R9 A0A0P6HG60 A0A2S2Q1D6 E2A629 A0A2S2NCV5 A0A1L3IWL4 A0A2H8TD64 J9K4C5 W5M8G4 A0A1B6BZG2 A0A310U8U7 A0A1B6CG97 A0A093H8L6 A0A1L8FLG8 A0A091ULW6 A0A3P9CK48 A0A093P3M7 A0A087RIS1
A0A158NYX8 A0A3L8DX23 V9ILI0 A0A1L8E4M1 A0A2A3EEV9 E2C0H1 F4W5H8 A0A088AN61 A0A026WAJ4 A0A151J3S1 A0A0N0U4H6 A0A0L7RDS6 K7IQL0 A0A0C9RP34 A0A067QKG8 A0A195BUM2 A0A0C9S320 A0A195D0X8 Q9VQY4 A0A2J7PQ37 A0A0Q5W964 B3N3Q6 A2RVF0 Q8MS50 A0A0J9QVL0 B4QA58 A0A154PC74 V5I802 D2A0Y3 Q9UAF0 A0A1J1IP34 A0A0R1DNL5 A0A1W4XW86 B4NYI9 B4I376 B3MLP2 A0A1W4XV81 A0A0P8XDZ5 A0A0T6BD08 A0A336LSN6 A0A336LS63 B4MDD2 A0A0Q9WLU5 B4N0N8 A0A0M4E9X6 A0A0U9HSJ3 A0A1Y1LFF6 A0A146LKB6 A0A146LYU6 A0A224X8K1 A0A023F379 A0A1B0CSL8 A0A0A9W0J1 A0A0P5U3B1 A0A0P6DCB0 A0A0P5I7T7 A0A0P5Q8Q9 A0A0P6G3P6 A0A0P6GAF5 A0A0P4YV08 E9HL04 A0A0P6F6A8 A0A0P5VQP2 A0A0A7ARN0 A0A0P5QQ66 A0A0P6AWT6 T1ITI8 A0A067QT28 A0A0P5X8I6 A0A232F3L7 A0A1S3IHP1 A0A2J7PQ84 K7J0G4 A0A2C9JEL2 A0A2C9JEL6 A0A2C9JEL7 A0A195D0R9 A0A0P6HG60 A0A2S2Q1D6 E2A629 A0A2S2NCV5 A0A1L3IWL4 A0A2H8TD64 J9K4C5 W5M8G4 A0A1B6BZG2 A0A310U8U7 A0A1B6CG97 A0A093H8L6 A0A1L8FLG8 A0A091ULW6 A0A3P9CK48 A0A093P3M7 A0A087RIS1
Pubmed
19121390
22118469
26354079
20798317
21347285
30249741
+ More
21719571 24508170 20075255 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 22936249 18362917 19820115 8950175 17550304 28004739 26823975 25474469 25401762 21292972 28648823 15562597 27762356 25186727
21719571 24508170 20075255 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 22936249 18362917 19820115 8950175 17550304 28004739 26823975 25474469 25401762 21292972 28648823 15562597 27762356 25186727
EMBL
BABH01019639
BABH01019640
ODYU01009789
SOQ54444.1
AGBW02012508
OWR44973.1
+ More
KQ459592 KPI96884.1 KQ460642 KPJ13322.1 GL437108 EFN71086.1 ADTU01004385 QOIP01000003 RLU24926.1 JR053625 AEY61978.1 GFDF01000525 JAV13559.1 KZ288269 PBC30014.1 GL451783 EFN78564.1 GL887645 EGI70628.1 KK107295 EZA53080.1 KQ980249 KYN17092.1 KQ435824 KOX72124.1 KQ414613 KOC69008.1 GBYB01015295 GBYB01015297 JAG85062.1 JAG85064.1 KK853235 KDR09575.1 KQ976407 KYM91447.1 GBYB01015296 JAG85063.1 KQ977004 KYN06511.1 AE014134 AAF51027.1 AAN10342.1 ABV53618.1 NEVH01022640 PNF18457.1 CH954177 KQS70086.1 EDV57715.1 BT029941 BT029944 ABM92815.1 ABM92818.1 ABV53617.1 AY119101 AAM50961.1 CM002910 KMY88046.1 CM000361 EDX03696.1 KMY88045.1 KQ434864 KZC09024.1 GALX01005540 JAB62926.1 KQ971338 EFA02575.1 AB030450 BAA83106.1 CVRI01000054 CRL00838.1 CM000157 KRJ97266.1 EDW87622.2 KRJ97265.1 CH480820 EDW54221.1 CH902620 EDV30763.1 KPU73027.1 LJIG01001728 KRT85212.1 UFQT01000144 SSX20845.1 SSX20846.1 CH940661 EDW71193.1 KRF85522.1 CH963920 EDW77651.2 CP012523 ALC37935.1 GARF01000010 JAC88927.1 GEZM01061808 JAV70306.1 GDHC01011333 JAQ07296.1 GDHC01005868 JAQ12761.1 GFTR01007729 JAW08697.1 GBBI01003086 JAC15626.1 AJWK01026236 AJWK01026237 GBHO01041657 JAG01947.1 GDIP01118779 JAL84935.1 GDIQ01079329 JAN15408.1 GDIP01148690 GDIQ01222046 LRGB01000141 JAJ74712.1 JAK29679.1 KZS20629.1 GDIQ01123836 JAL27890.1 GDIQ01038872 JAN55865.1 GDIQ01038871 JAN55866.1 GDIP01221831 JAJ01571.1 GL732674 EFX67564.1 GDIQ01064899 JAN29838.1 GDIP01096852 JAM06863.1 KF906302 AHK05667.1 GDIQ01117534 JAL34192.1 GDIP01023214 JAM80501.1 JH431485 KK852972 KDR13078.1 GDIP01075833 JAM27882.1 NNAY01001049 OXU25341.1 PNF18493.1 KYN06510.1 GDIQ01019922 JAN74815.1 GGMS01002375 MBY71578.1 EFN71088.1 GGMR01002391 MBY15010.1 KX056437 APG57147.1 GFXV01000246 MBW12051.1 ABLF02036674 AHAT01024180 GEDC01030646 JAS06652.1 KV467275 OCT56466.1 GEDC01030780 GEDC01024857 GEDC01012812 GEDC01006698 GEDC01001329 JAS06518.1 JAS12441.1 JAS24486.1 JAS30600.1 JAS35969.1 KL206095 KFV78918.1 CM004478 OCT72417.1 KK457451 KFQ77156.1 KL225327 KFW71006.1 KL226389 KFM13375.1
KQ459592 KPI96884.1 KQ460642 KPJ13322.1 GL437108 EFN71086.1 ADTU01004385 QOIP01000003 RLU24926.1 JR053625 AEY61978.1 GFDF01000525 JAV13559.1 KZ288269 PBC30014.1 GL451783 EFN78564.1 GL887645 EGI70628.1 KK107295 EZA53080.1 KQ980249 KYN17092.1 KQ435824 KOX72124.1 KQ414613 KOC69008.1 GBYB01015295 GBYB01015297 JAG85062.1 JAG85064.1 KK853235 KDR09575.1 KQ976407 KYM91447.1 GBYB01015296 JAG85063.1 KQ977004 KYN06511.1 AE014134 AAF51027.1 AAN10342.1 ABV53618.1 NEVH01022640 PNF18457.1 CH954177 KQS70086.1 EDV57715.1 BT029941 BT029944 ABM92815.1 ABM92818.1 ABV53617.1 AY119101 AAM50961.1 CM002910 KMY88046.1 CM000361 EDX03696.1 KMY88045.1 KQ434864 KZC09024.1 GALX01005540 JAB62926.1 KQ971338 EFA02575.1 AB030450 BAA83106.1 CVRI01000054 CRL00838.1 CM000157 KRJ97266.1 EDW87622.2 KRJ97265.1 CH480820 EDW54221.1 CH902620 EDV30763.1 KPU73027.1 LJIG01001728 KRT85212.1 UFQT01000144 SSX20845.1 SSX20846.1 CH940661 EDW71193.1 KRF85522.1 CH963920 EDW77651.2 CP012523 ALC37935.1 GARF01000010 JAC88927.1 GEZM01061808 JAV70306.1 GDHC01011333 JAQ07296.1 GDHC01005868 JAQ12761.1 GFTR01007729 JAW08697.1 GBBI01003086 JAC15626.1 AJWK01026236 AJWK01026237 GBHO01041657 JAG01947.1 GDIP01118779 JAL84935.1 GDIQ01079329 JAN15408.1 GDIP01148690 GDIQ01222046 LRGB01000141 JAJ74712.1 JAK29679.1 KZS20629.1 GDIQ01123836 JAL27890.1 GDIQ01038872 JAN55865.1 GDIQ01038871 JAN55866.1 GDIP01221831 JAJ01571.1 GL732674 EFX67564.1 GDIQ01064899 JAN29838.1 GDIP01096852 JAM06863.1 KF906302 AHK05667.1 GDIQ01117534 JAL34192.1 GDIP01023214 JAM80501.1 JH431485 KK852972 KDR13078.1 GDIP01075833 JAM27882.1 NNAY01001049 OXU25341.1 PNF18493.1 KYN06510.1 GDIQ01019922 JAN74815.1 GGMS01002375 MBY71578.1 EFN71088.1 GGMR01002391 MBY15010.1 KX056437 APG57147.1 GFXV01000246 MBW12051.1 ABLF02036674 AHAT01024180 GEDC01030646 JAS06652.1 KV467275 OCT56466.1 GEDC01030780 GEDC01024857 GEDC01012812 GEDC01006698 GEDC01001329 JAS06518.1 JAS12441.1 JAS24486.1 JAS30600.1 JAS35969.1 KL206095 KFV78918.1 CM004478 OCT72417.1 KK457451 KFQ77156.1 KL225327 KFW71006.1 KL226389 KFM13375.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000000311
UP000005205
+ More
UP000279307 UP000242457 UP000008237 UP000007755 UP000005203 UP000053097 UP000078492 UP000053105 UP000053825 UP000002358 UP000027135 UP000078540 UP000078542 UP000000803 UP000235965 UP000008711 UP000000304 UP000076502 UP000007266 UP000183832 UP000002282 UP000192223 UP000001292 UP000007801 UP000008792 UP000007798 UP000092553 UP000092461 UP000076858 UP000000305 UP000215335 UP000085678 UP000076420 UP000007819 UP000018468 UP000053584 UP000186698 UP000265160 UP000054081 UP000053286
UP000279307 UP000242457 UP000008237 UP000007755 UP000005203 UP000053097 UP000078492 UP000053105 UP000053825 UP000002358 UP000027135 UP000078540 UP000078542 UP000000803 UP000235965 UP000008711 UP000000304 UP000076502 UP000007266 UP000183832 UP000002282 UP000192223 UP000001292 UP000007801 UP000008792 UP000007798 UP000092553 UP000092461 UP000076858 UP000000305 UP000215335 UP000085678 UP000076420 UP000007819 UP000018468 UP000053584 UP000186698 UP000265160 UP000054081 UP000053286
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
H9J6F2
A0A2H1WPU0
A0A212EU03
A0A194PVU7
A0A0N0PC93
E2A627
+ More
A0A158NYX8 A0A3L8DX23 V9ILI0 A0A1L8E4M1 A0A2A3EEV9 E2C0H1 F4W5H8 A0A088AN61 A0A026WAJ4 A0A151J3S1 A0A0N0U4H6 A0A0L7RDS6 K7IQL0 A0A0C9RP34 A0A067QKG8 A0A195BUM2 A0A0C9S320 A0A195D0X8 Q9VQY4 A0A2J7PQ37 A0A0Q5W964 B3N3Q6 A2RVF0 Q8MS50 A0A0J9QVL0 B4QA58 A0A154PC74 V5I802 D2A0Y3 Q9UAF0 A0A1J1IP34 A0A0R1DNL5 A0A1W4XW86 B4NYI9 B4I376 B3MLP2 A0A1W4XV81 A0A0P8XDZ5 A0A0T6BD08 A0A336LSN6 A0A336LS63 B4MDD2 A0A0Q9WLU5 B4N0N8 A0A0M4E9X6 A0A0U9HSJ3 A0A1Y1LFF6 A0A146LKB6 A0A146LYU6 A0A224X8K1 A0A023F379 A0A1B0CSL8 A0A0A9W0J1 A0A0P5U3B1 A0A0P6DCB0 A0A0P5I7T7 A0A0P5Q8Q9 A0A0P6G3P6 A0A0P6GAF5 A0A0P4YV08 E9HL04 A0A0P6F6A8 A0A0P5VQP2 A0A0A7ARN0 A0A0P5QQ66 A0A0P6AWT6 T1ITI8 A0A067QT28 A0A0P5X8I6 A0A232F3L7 A0A1S3IHP1 A0A2J7PQ84 K7J0G4 A0A2C9JEL2 A0A2C9JEL6 A0A2C9JEL7 A0A195D0R9 A0A0P6HG60 A0A2S2Q1D6 E2A629 A0A2S2NCV5 A0A1L3IWL4 A0A2H8TD64 J9K4C5 W5M8G4 A0A1B6BZG2 A0A310U8U7 A0A1B6CG97 A0A093H8L6 A0A1L8FLG8 A0A091ULW6 A0A3P9CK48 A0A093P3M7 A0A087RIS1
A0A158NYX8 A0A3L8DX23 V9ILI0 A0A1L8E4M1 A0A2A3EEV9 E2C0H1 F4W5H8 A0A088AN61 A0A026WAJ4 A0A151J3S1 A0A0N0U4H6 A0A0L7RDS6 K7IQL0 A0A0C9RP34 A0A067QKG8 A0A195BUM2 A0A0C9S320 A0A195D0X8 Q9VQY4 A0A2J7PQ37 A0A0Q5W964 B3N3Q6 A2RVF0 Q8MS50 A0A0J9QVL0 B4QA58 A0A154PC74 V5I802 D2A0Y3 Q9UAF0 A0A1J1IP34 A0A0R1DNL5 A0A1W4XW86 B4NYI9 B4I376 B3MLP2 A0A1W4XV81 A0A0P8XDZ5 A0A0T6BD08 A0A336LSN6 A0A336LS63 B4MDD2 A0A0Q9WLU5 B4N0N8 A0A0M4E9X6 A0A0U9HSJ3 A0A1Y1LFF6 A0A146LKB6 A0A146LYU6 A0A224X8K1 A0A023F379 A0A1B0CSL8 A0A0A9W0J1 A0A0P5U3B1 A0A0P6DCB0 A0A0P5I7T7 A0A0P5Q8Q9 A0A0P6G3P6 A0A0P6GAF5 A0A0P4YV08 E9HL04 A0A0P6F6A8 A0A0P5VQP2 A0A0A7ARN0 A0A0P5QQ66 A0A0P6AWT6 T1ITI8 A0A067QT28 A0A0P5X8I6 A0A232F3L7 A0A1S3IHP1 A0A2J7PQ84 K7J0G4 A0A2C9JEL2 A0A2C9JEL6 A0A2C9JEL7 A0A195D0R9 A0A0P6HG60 A0A2S2Q1D6 E2A629 A0A2S2NCV5 A0A1L3IWL4 A0A2H8TD64 J9K4C5 W5M8G4 A0A1B6BZG2 A0A310U8U7 A0A1B6CG97 A0A093H8L6 A0A1L8FLG8 A0A091ULW6 A0A3P9CK48 A0A093P3M7 A0A087RIS1
PDB
6FFC
E-value=8.30703e-48,
Score=483
Ontologies
GO
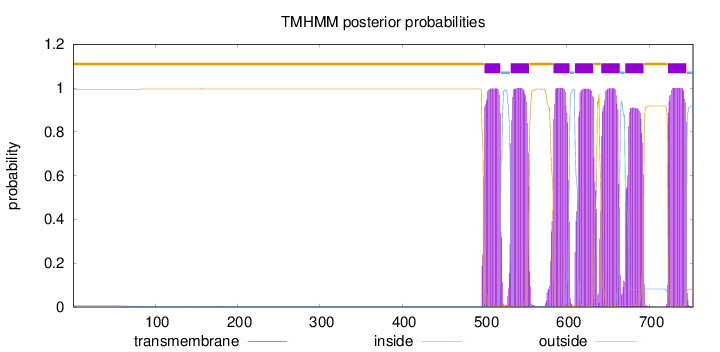
Topology
Length:
753
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
150.54153
Exp number, first 60 AAs:
0.00023
Total prob of N-in:
0.00520
outside
1 - 499
TMhelix
500 - 519
inside
520 - 531
TMhelix
532 - 554
outside
555 - 583
TMhelix
584 - 603
inside
604 - 609
TMhelix
610 - 632
outside
633 - 641
TMhelix
642 - 664
inside
665 - 670
TMhelix
671 - 693
outside
694 - 722
TMhelix
723 - 745
inside
746 - 753
Population Genetic Test Statistics
Pi
30.481674
Theta
25.243064
Tajima's D
0.458034
CLR
0.6223
CSRT
0.513574321283936
Interpretation
Uncertain
