Pre Gene Modal
BGIBMGA005888
Annotation
PREDICTED:_ELAV-like_protein_2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.564
Sequence
CDS
ATGATGGCAAATCCGCTCGACTCTCTAAACCAGCCCACGCAAAATGGAAGTAAGTTGCAGCCGTGCAAGAATGAGTCTAAGACGAATTTGATAATCAATTATCTGCCGCAAACGATGACCCAGGAGGAAATTCGATCATTATTTTCGAGTGTCGGCGAAGTAGAAAGCTGCAAGTTAATAAGGGACAAGGTGACCGTGTTTCCGGATCATATTCTCAACGGTCAGAGTCTTGGATACGCTTTCGTCAACTATCACCGAGCTGAGGATGCGGAAAAGGCGGTGAATACACTCAATGGTCTCAGACTGCAGAACAAGATCATCAAAGTTTCATACGCACGTCCAAGCTCTGATGCGATTAAAGGTGCCAACCTCTACGTCTCCGGACTCCCGAAACACATGACTCAGCAAGAATTGGAGAAACTCTTCGGCCCCTACGGTACCATTATCAGCTCGCGCATTCTCCATGAGAACATGAATGTCGGTCAGCTGATGCAGGTAGCTCCAGAAGATCACTCTATCCAAGGTCCATCAAGAGGAGTCGCCTTCATCCGGTACGATCAAAGGTGTGAAGCAGAAGCAGCTATACGTGAGTTGAATGGCACTATTCCACCTGGTGGAACAACCCCAATGACTGTCAAGTGTGCAAACAACCCAAGCAATCAGAATAAGGTTTTGGCACCCCTTACTGCATACCTGGCTCCAACTTCAACACGGCGCATCATAACCCCTGCAGGTGGATCAGGCTGGTGTATCTTCGTGTACAACATCGGGGCTGACACTGAGGAGAGTGTTCTGTGGCAACTTTTTGGGCCATTTGGTGCCGTCCAGAGTGTGAAAATAATTAGAGATCCCTCTACAAACAAGTGCAAAGGTTATGGTTTTGTTACTATGACCAATTATGATGAGGCTGTTGTTGCCATTCAATCCCTGAATGGCTATTCACTGAACGGTCAGGTCCTTCAGGTCAGCTTCAAGACCAACAAGAGTAAATCTTAA
Protein
MMANPLDSLNQPTQNGSKLQPCKNESKTNLIINYLPQTMTQEEIRSLFSSVGEVESCKLIRDKVTVFPDHILNGQSLGYAFVNYHRAEDAEKAVNTLNGLRLQNKIIKVSYARPSSDAIKGANLYVSGLPKHMTQQELEKLFGPYGTIISSRILHENMNVGQLMQVAPEDHSIQGPSRGVAFIRYDQRCEAEAAIRELNGTIPPGGTTPMTVKCANNPSNQNKVLAPLTAYLAPTSTRRIITPAGGSGWCIFVYNIGADTEESVLWQLFGPFGAVQSVKIIRDPSTNKCKGYGFVTMTNYDEAVVAIQSLNGYSLNGQVLQVSFKTNKSKS
Summary
Uniprot
A0A0N0PBM7
A0A194PV67
H9J8P6
A0A2A4J1E6
A0A2H1W891
I4DPM2
+ More
A0A212F563 A0A1Y1NCP8 A0A0T6BAV5 A0A1Y1NCR0 A0A139WPC8 V9I8A0 A0A1B0CNX6 N6T223 U4UME0 A0A1B6DYH7 A0A1B6FAB4 A0A1B6FNG9 A0A1B6FMI4 A0A1B6HCD8 V9I7T6 A0A1B6IE92 A0A139WPK5 A0A1B6MPA0 A0A2A3EDH0 A0A1B6L213 A0A1B6BYP2 A0A1B6LZ91 A0A1B6IH54 A0A1B6HLT5 A0A1B6L4M1 A0A1B6JSA0 A0A1B6KIN8 A0A0L7LF28 A0A1B6LLS9 A0A1B6M3J1 A0A1Q3FNL0 A0A0A9WML8 B0W031 A0A0A9WPM1 A0A0K8T382 B3MS25 A0A0K8T3U4 B0X9Z1 B4L604 A0A1W4VJY2 B4M748 B4Q1Z0 A0A0M4EU27 B4NEI3 B4JM01 B3NX84 E1NZB4 A0A3B0KLA2 B5DNV4 A0A146KP32 A0A146MFB4 W8AKC4 E0VBM6 A0A034V4J7 A0A0N8NZT3 E9IGR2 A0A0Q9WVT0 A0A1W4VKQ0 A0A0Q5TKR8 Q9VYI0 A0A0R1EAH8 A0A0Q9XEN8 A0A3B0KRV0 A0A0R3P1Z1 A0A1B0D1N4 A0A0A1XJ57 A0A0A1WSN6 A0A1I8MKP7 A0A1I8Q1M4 A0A1I8MKP3 B4IGK8 A0A1I8Q1K5 A0A1B0G809 A0A1B0AY01 T1K3Z8 A0A1A9VWP4 A0A1L8DGL4 A0A1A9WWW7 A0A1J1HR36 A0A2L2Y0N9 A0A1L8DH41 A0A1L8DH01 A0A2R7WD77 U5ER87 A0A336LT12 A0A248XBD7 A6YB86 A0A1B0GQI6 C3YCD7 Q966S6 A0A223FSJ8 Q7YTD6 A0A182KM55 Q7PYD4
A0A212F563 A0A1Y1NCP8 A0A0T6BAV5 A0A1Y1NCR0 A0A139WPC8 V9I8A0 A0A1B0CNX6 N6T223 U4UME0 A0A1B6DYH7 A0A1B6FAB4 A0A1B6FNG9 A0A1B6FMI4 A0A1B6HCD8 V9I7T6 A0A1B6IE92 A0A139WPK5 A0A1B6MPA0 A0A2A3EDH0 A0A1B6L213 A0A1B6BYP2 A0A1B6LZ91 A0A1B6IH54 A0A1B6HLT5 A0A1B6L4M1 A0A1B6JSA0 A0A1B6KIN8 A0A0L7LF28 A0A1B6LLS9 A0A1B6M3J1 A0A1Q3FNL0 A0A0A9WML8 B0W031 A0A0A9WPM1 A0A0K8T382 B3MS25 A0A0K8T3U4 B0X9Z1 B4L604 A0A1W4VJY2 B4M748 B4Q1Z0 A0A0M4EU27 B4NEI3 B4JM01 B3NX84 E1NZB4 A0A3B0KLA2 B5DNV4 A0A146KP32 A0A146MFB4 W8AKC4 E0VBM6 A0A034V4J7 A0A0N8NZT3 E9IGR2 A0A0Q9WVT0 A0A1W4VKQ0 A0A0Q5TKR8 Q9VYI0 A0A0R1EAH8 A0A0Q9XEN8 A0A3B0KRV0 A0A0R3P1Z1 A0A1B0D1N4 A0A0A1XJ57 A0A0A1WSN6 A0A1I8MKP7 A0A1I8Q1M4 A0A1I8MKP3 B4IGK8 A0A1I8Q1K5 A0A1B0G809 A0A1B0AY01 T1K3Z8 A0A1A9VWP4 A0A1L8DGL4 A0A1A9WWW7 A0A1J1HR36 A0A2L2Y0N9 A0A1L8DH41 A0A1L8DH01 A0A2R7WD77 U5ER87 A0A336LT12 A0A248XBD7 A6YB86 A0A1B0GQI6 C3YCD7 Q966S6 A0A223FSJ8 Q7YTD6 A0A182KM55 Q7PYD4
Pubmed
26354079
19121390
22651552
22118469
28004739
18362917
+ More
19820115 23537049 26227816 25401762 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 26823975 24495485 20566863 25348373 21282665 18057021 12324951 12591606 25830018 25315136 26561354 17448990 18563158 11754014 28765531 12837244 20966253 12364791
19820115 23537049 26227816 25401762 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 26823975 24495485 20566863 25348373 21282665 18057021 12324951 12591606 25830018 25315136 26561354 17448990 18563158 11754014 28765531 12837244 20966253 12364791
EMBL
KQ460954
KPJ10266.1
KQ459592
KPI96883.1
BABH01030258
NWSH01004215
+ More
PCG65344.1 ODYU01006980 SOQ49315.1 AK403674 BAM19862.1 AGBW02010238 OWR48876.1 GEZM01006631 JAV95671.1 LJIG01002450 KRT84490.1 GEZM01006629 JAV95673.1 KQ971307 KYB29716.1 JR035794 JR035797 JR035798 JR035799 AEY56686.1 AEY56689.1 AEY56690.1 AEY56691.1 AJWK01021069 AJWK01021070 APGK01047133 APGK01047134 KB741077 ENN74149.1 KB632384 ERL94282.1 GEDC01021269 GEDC01006570 JAS16029.1 JAS30728.1 GECZ01022590 JAS47179.1 GECZ01018021 JAS51748.1 GECZ01018343 JAS51426.1 GECU01035349 GECU01017493 JAS72357.1 JAS90213.1 JR035802 JR035803 JR035804 JR035805 AEY56694.1 AEY56695.1 AEY56696.1 AEY56697.1 GECU01022448 JAS85258.1 KYB29715.1 GEBQ01002266 JAT37711.1 KZ288291 PBC29216.1 GEBQ01022296 JAT17681.1 GEDC01030930 GEDC01027761 JAS06368.1 JAS09537.1 GEBQ01011043 JAT28934.1 GECU01021467 JAS86239.1 GECU01032046 JAS75660.1 GEBQ01021308 JAT18669.1 GECU01005704 JAT02003.1 GEBQ01028657 JAT11320.1 JTDY01001464 KOB73801.1 GEBQ01015361 JAT24616.1 GEBQ01009458 JAT30519.1 GFDL01005856 JAV29189.1 GBHO01033902 JAG09702.1 DS231816 EDS37603.1 GBHO01033905 JAG09699.1 GBRD01005965 JAG59856.1 CH902622 EDV34580.1 GBRD01005966 JAG59855.1 DS232556 EDS43417.1 CH933812 EDW05800.1 CH940653 EDW62615.1 CM000162 EDX01511.1 CP012528 ALC48510.1 CH964239 EDW82152.1 CH916371 EDV91762.1 CH954180 EDV47256.1 BT125663 AE014298 ADN43900.1 AFH07361.1 AFH07363.2 AHN59643.1 OUUW01000021 SPP89390.1 CH379065 EDY73206.1 GDHC01020368 JAP98260.1 GDHC01001339 JAQ17290.1 GAMC01017485 GAMC01017483 JAB89070.1 DS235033 EEB10782.1 GAKP01021503 GAKP01021499 JAC37449.1 KPU75204.1 GL763054 EFZ20185.1 KRF99576.1 KQS30422.1 KQS30423.1 AF239667 BT120384 AAF43091.1 AAF48215.3 ABW09403.1 ADD16464.1 AFH07360.1 AFH07362.1 AGB95339.1 AGB95340.1 KRK06190.1 KRK06191.1 KRG07039.1 SPP89389.1 KRT07226.1 AJVK01010341 AJVK01010342 GBXI01003305 JAD10987.1 GBXI01012767 JAD01525.1 CH480836 EDW48958.1 CCAG010000149 CCAG010000150 JXJN01005475 JXJN01005476 CAEY01001563 GFDF01008513 JAV05571.1 CVRI01000014 CRK89852.1 IAAA01006536 LAA01731.1 GFDF01008420 JAV05664.1 GFDF01008419 JAV05665.1 KK854488 PTY16275.1 GANO01003851 JAB56020.1 UFQT01000095 SSX19729.1 KY569299 ASW25830.1 EF384209 ABO93208.1 AJVK01007258 AJVK01007259 GG666501 EEN61974.1 AB066563 BAB62225.1 MF409251 AST12253.1 AY313137 AAP79277.1 AAAB01008987 EAA01735.5
PCG65344.1 ODYU01006980 SOQ49315.1 AK403674 BAM19862.1 AGBW02010238 OWR48876.1 GEZM01006631 JAV95671.1 LJIG01002450 KRT84490.1 GEZM01006629 JAV95673.1 KQ971307 KYB29716.1 JR035794 JR035797 JR035798 JR035799 AEY56686.1 AEY56689.1 AEY56690.1 AEY56691.1 AJWK01021069 AJWK01021070 APGK01047133 APGK01047134 KB741077 ENN74149.1 KB632384 ERL94282.1 GEDC01021269 GEDC01006570 JAS16029.1 JAS30728.1 GECZ01022590 JAS47179.1 GECZ01018021 JAS51748.1 GECZ01018343 JAS51426.1 GECU01035349 GECU01017493 JAS72357.1 JAS90213.1 JR035802 JR035803 JR035804 JR035805 AEY56694.1 AEY56695.1 AEY56696.1 AEY56697.1 GECU01022448 JAS85258.1 KYB29715.1 GEBQ01002266 JAT37711.1 KZ288291 PBC29216.1 GEBQ01022296 JAT17681.1 GEDC01030930 GEDC01027761 JAS06368.1 JAS09537.1 GEBQ01011043 JAT28934.1 GECU01021467 JAS86239.1 GECU01032046 JAS75660.1 GEBQ01021308 JAT18669.1 GECU01005704 JAT02003.1 GEBQ01028657 JAT11320.1 JTDY01001464 KOB73801.1 GEBQ01015361 JAT24616.1 GEBQ01009458 JAT30519.1 GFDL01005856 JAV29189.1 GBHO01033902 JAG09702.1 DS231816 EDS37603.1 GBHO01033905 JAG09699.1 GBRD01005965 JAG59856.1 CH902622 EDV34580.1 GBRD01005966 JAG59855.1 DS232556 EDS43417.1 CH933812 EDW05800.1 CH940653 EDW62615.1 CM000162 EDX01511.1 CP012528 ALC48510.1 CH964239 EDW82152.1 CH916371 EDV91762.1 CH954180 EDV47256.1 BT125663 AE014298 ADN43900.1 AFH07361.1 AFH07363.2 AHN59643.1 OUUW01000021 SPP89390.1 CH379065 EDY73206.1 GDHC01020368 JAP98260.1 GDHC01001339 JAQ17290.1 GAMC01017485 GAMC01017483 JAB89070.1 DS235033 EEB10782.1 GAKP01021503 GAKP01021499 JAC37449.1 KPU75204.1 GL763054 EFZ20185.1 KRF99576.1 KQS30422.1 KQS30423.1 AF239667 BT120384 AAF43091.1 AAF48215.3 ABW09403.1 ADD16464.1 AFH07360.1 AFH07362.1 AGB95339.1 AGB95340.1 KRK06190.1 KRK06191.1 KRG07039.1 SPP89389.1 KRT07226.1 AJVK01010341 AJVK01010342 GBXI01003305 JAD10987.1 GBXI01012767 JAD01525.1 CH480836 EDW48958.1 CCAG010000149 CCAG010000150 JXJN01005475 JXJN01005476 CAEY01001563 GFDF01008513 JAV05571.1 CVRI01000014 CRK89852.1 IAAA01006536 LAA01731.1 GFDF01008420 JAV05664.1 GFDF01008419 JAV05665.1 KK854488 PTY16275.1 GANO01003851 JAB56020.1 UFQT01000095 SSX19729.1 KY569299 ASW25830.1 EF384209 ABO93208.1 AJVK01007258 AJVK01007259 GG666501 EEN61974.1 AB066563 BAB62225.1 MF409251 AST12253.1 AY313137 AAP79277.1 AAAB01008987 EAA01735.5
Proteomes
UP000053240
UP000053268
UP000005204
UP000218220
UP000007151
UP000007266
+ More
UP000092461 UP000019118 UP000030742 UP000242457 UP000037510 UP000002320 UP000007801 UP000009192 UP000192221 UP000008792 UP000002282 UP000092553 UP000007798 UP000001070 UP000008711 UP000000803 UP000268350 UP000001819 UP000009046 UP000092462 UP000095301 UP000095300 UP000001292 UP000092444 UP000092460 UP000015104 UP000078200 UP000091820 UP000183832 UP000001554 UP000075882 UP000007062
UP000092461 UP000019118 UP000030742 UP000242457 UP000037510 UP000002320 UP000007801 UP000009192 UP000192221 UP000008792 UP000002282 UP000092553 UP000007798 UP000001070 UP000008711 UP000000803 UP000268350 UP000001819 UP000009046 UP000092462 UP000095301 UP000095300 UP000001292 UP000092444 UP000092460 UP000015104 UP000078200 UP000091820 UP000183832 UP000001554 UP000075882 UP000007062
Pfam
PF00076 RRM_1
Interpro
SUPFAM
SSF54928
SSF54928
Gene 3D
CDD
ProteinModelPortal
A0A0N0PBM7
A0A194PV67
H9J8P6
A0A2A4J1E6
A0A2H1W891
I4DPM2
+ More
A0A212F563 A0A1Y1NCP8 A0A0T6BAV5 A0A1Y1NCR0 A0A139WPC8 V9I8A0 A0A1B0CNX6 N6T223 U4UME0 A0A1B6DYH7 A0A1B6FAB4 A0A1B6FNG9 A0A1B6FMI4 A0A1B6HCD8 V9I7T6 A0A1B6IE92 A0A139WPK5 A0A1B6MPA0 A0A2A3EDH0 A0A1B6L213 A0A1B6BYP2 A0A1B6LZ91 A0A1B6IH54 A0A1B6HLT5 A0A1B6L4M1 A0A1B6JSA0 A0A1B6KIN8 A0A0L7LF28 A0A1B6LLS9 A0A1B6M3J1 A0A1Q3FNL0 A0A0A9WML8 B0W031 A0A0A9WPM1 A0A0K8T382 B3MS25 A0A0K8T3U4 B0X9Z1 B4L604 A0A1W4VJY2 B4M748 B4Q1Z0 A0A0M4EU27 B4NEI3 B4JM01 B3NX84 E1NZB4 A0A3B0KLA2 B5DNV4 A0A146KP32 A0A146MFB4 W8AKC4 E0VBM6 A0A034V4J7 A0A0N8NZT3 E9IGR2 A0A0Q9WVT0 A0A1W4VKQ0 A0A0Q5TKR8 Q9VYI0 A0A0R1EAH8 A0A0Q9XEN8 A0A3B0KRV0 A0A0R3P1Z1 A0A1B0D1N4 A0A0A1XJ57 A0A0A1WSN6 A0A1I8MKP7 A0A1I8Q1M4 A0A1I8MKP3 B4IGK8 A0A1I8Q1K5 A0A1B0G809 A0A1B0AY01 T1K3Z8 A0A1A9VWP4 A0A1L8DGL4 A0A1A9WWW7 A0A1J1HR36 A0A2L2Y0N9 A0A1L8DH41 A0A1L8DH01 A0A2R7WD77 U5ER87 A0A336LT12 A0A248XBD7 A6YB86 A0A1B0GQI6 C3YCD7 Q966S6 A0A223FSJ8 Q7YTD6 A0A182KM55 Q7PYD4
A0A212F563 A0A1Y1NCP8 A0A0T6BAV5 A0A1Y1NCR0 A0A139WPC8 V9I8A0 A0A1B0CNX6 N6T223 U4UME0 A0A1B6DYH7 A0A1B6FAB4 A0A1B6FNG9 A0A1B6FMI4 A0A1B6HCD8 V9I7T6 A0A1B6IE92 A0A139WPK5 A0A1B6MPA0 A0A2A3EDH0 A0A1B6L213 A0A1B6BYP2 A0A1B6LZ91 A0A1B6IH54 A0A1B6HLT5 A0A1B6L4M1 A0A1B6JSA0 A0A1B6KIN8 A0A0L7LF28 A0A1B6LLS9 A0A1B6M3J1 A0A1Q3FNL0 A0A0A9WML8 B0W031 A0A0A9WPM1 A0A0K8T382 B3MS25 A0A0K8T3U4 B0X9Z1 B4L604 A0A1W4VJY2 B4M748 B4Q1Z0 A0A0M4EU27 B4NEI3 B4JM01 B3NX84 E1NZB4 A0A3B0KLA2 B5DNV4 A0A146KP32 A0A146MFB4 W8AKC4 E0VBM6 A0A034V4J7 A0A0N8NZT3 E9IGR2 A0A0Q9WVT0 A0A1W4VKQ0 A0A0Q5TKR8 Q9VYI0 A0A0R1EAH8 A0A0Q9XEN8 A0A3B0KRV0 A0A0R3P1Z1 A0A1B0D1N4 A0A0A1XJ57 A0A0A1WSN6 A0A1I8MKP7 A0A1I8Q1M4 A0A1I8MKP3 B4IGK8 A0A1I8Q1K5 A0A1B0G809 A0A1B0AY01 T1K3Z8 A0A1A9VWP4 A0A1L8DGL4 A0A1A9WWW7 A0A1J1HR36 A0A2L2Y0N9 A0A1L8DH41 A0A1L8DH01 A0A2R7WD77 U5ER87 A0A336LT12 A0A248XBD7 A6YB86 A0A1B0GQI6 C3YCD7 Q966S6 A0A223FSJ8 Q7YTD6 A0A182KM55 Q7PYD4
PDB
1G2E
E-value=1.37588e-54,
Score=538
Ontologies
GO
Topology
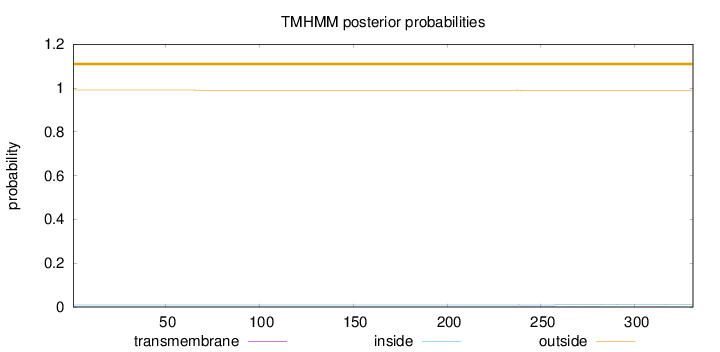
Length:
331
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04684
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00951
outside
1 - 331
Population Genetic Test Statistics
Pi
6.118332
Theta
6.401791
Tajima's D
-0.125255
CLR
1.946029
CSRT
0.355582220888956
Interpretation
Uncertain
