Pre Gene Modal
BGIBMGA005889
Annotation
PREDICTED:_GRIP_and_coiled-coil_domain-containing_protein_1_isoform_X2_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.896 Cytoskeletal Reliability : 1.625
Sequence
CDS
ATGGAATCATTATCAAAACAAGAGCTAATATCTACCATAACAAAACAAGCTGATCAGATAAAACGATACGAATCACGTTTACGGGATGTAGTCACAGCTTACAAGGGCTTGGTTAAAGAGAAAGAAGCATTAGAGATCAGCTTGAAGGCACTCAACAAGACTGAAAACACTATAGATGGAAGTGAAGATGATTCGGTCACAGCCCTCACTCTGTCTCTTTCTACCCTCACAGTGGAGAAAGCTCGCATGGAAGAAGCTTTCCAGGCAGACAAAAAAGTTACAAGGGATAAGTATGAGAACATGTTAGCGGCACTGAGAGAGGAGAATAAAATGCTATCGCAGCAACATTTGCTGGAGCTTAACAGTCTGAAAGCCAAGATAGCCTACGAGATACAAGAGAGGGAGAAGGAGAGAGCTGATCATGTTGCTATGATGAAAGAGTTGCATTTGAAATTGAACAGCGAAAGGAAGACGAAAGAGAAATTAGAAGACCGAGTAGTTAATTCAACTGAAGCTCAAGCGTCACAAGCCGAGCTAGAGAAGCGAGTTAGAGATCTGAGCAGTTCCTTGGAGGCAGCCCAAAGGAGGCTGCAGCGTGCCGAAGCCCGGACTGTTGAGACTCCAGCCCAATTAGTCAGGCTTCAGCAAAAATTGGCGCTGATCGAGCAATCACATTCAGTCGCTATCAGAGAAGAACAAATAAAAGCGAAAAGGGCTGAAGAATCTGCCAGGAAGATCTGTGCCCGTCAAGAAGAAAGAGTCGCGCTGCTAGAAGGAAAACTAGCTGAACTCTCAGAGACTATAGGACAGTACGATTTGAGGAGACGCCGCGACCAACAGCTGATACAGCAACTTAGAGATTCTATAAATGGGAGGCTGCTTGAGAAGATCGCTCCGGCTACGATTGAAGAAAACACTCCGGTCCAACTTAGCGAAACGGACAACGAGAAGTTGGCGGACAGCGAATATCTACAAACGTTAATCGACAAAATACACATCCTGAAGAAGGAATTGATATCGGAGAACGACAAGCAGGGCTCGCCCATAGACGTCGCCGCCGTGTTCCAAATCGAAGGGTTCGGTGACGTTCATCTCAAGTGCAGGGAGGATTACGAGAAGCTGCGTCTGGAGTTGGAGGAGGAGCGAGCACGTAGCTCCAGGGAGGGCGTCGAACCGCGACCCTCCTCGCCAGCTCTGCGCCACGACCTCGCGCTGCTCGCCGACCAGCTGCACACCTGCAAGCTGATGCTGGACGAGGAGAAGCAAGACAAAATCGACGCCCTGAGGATCTGCGAGGAGAAATTGAAAAATGAAGAGGAATACCACAGGGAGGTAACGACGGACCTGAAGAACAGGATCCACGTCCTCGAGAAGCAGGTCGCGACGCAGAGGGAGAGGTACGCCGCGCTGCTGGACGAGACGGACAGCTACATCCGCGGCAGGAACGAGCGGAAGGTCAGCAATGAGGAACACAACGTGTTTGCTGATGTGTCAGCGCCTCCCCACATGTTGCATTACGCCCACGAGCTCGCCCGGAAGGACCAGGACATATCGCAGCTCAGGAAGGACAAGCATCTGCTCGAGGGCCAGTACAGAGACTGCCAACGAGACGCGACCATAGAGAAAGAGCGCTTCAAAGAAGTCGTCAGAGCGCTGAAAGAGGAGATTGACAGATTAAGAAGAACGCAATCCCGTGAAGGTGCAAATTTAGAGTATTTGAAGAACGTTGTGCTCGCTTACTTGATGTCCACGGACTACGCCGGCAAGAGGCACATGCTAAACGCTATAGCCGCCGTGTTGCATTTCACTAACGCCGAACGTCACTCCGTGCTGTCTACTTTGTAA
Protein
MESLSKQELISTITKQADQIKRYESRLRDVVTAYKGLVKEKEALEISLKALNKTENTIDGSEDDSVTALTLSLSTLTVEKARMEEAFQADKKVTRDKYENMLAALREENKMLSQQHLLELNSLKAKIAYEIQEREKERADHVAMMKELHLKLNSERKTKEKLEDRVVNSTEAQASQAELEKRVRDLSSSLEAAQRRLQRAEARTVETPAQLVRLQQKLALIEQSHSVAIREEQIKAKRAEESARKICARQEERVALLEGKLAELSETIGQYDLRRRRDQQLIQQLRDSINGRLLEKIAPATIEENTPVQLSETDNEKLADSEYLQTLIDKIHILKKELISENDKQGSPIDVAAVFQIEGFGDVHLKCREDYEKLRLELEEERARSSREGVEPRPSSPALRHDLALLADQLHTCKLMLDEEKQDKIDALRICEEKLKNEEEYHREVTTDLKNRIHVLEKQVATQRERYAALLDETDSYIRGRNERKVSNEEHNVFADVSAPPHMLHYAHELARKDQDISQLRKDKHLLEGQYRDCQRDATIEKERFKEVVRALKEEIDRLRRTQSREGANLEYLKNVVLAYLMSTDYAGKRHMLNAIAAVLHFTNAERHSVLSTL
Summary
Uniprot
H9J8P7
A0A3S2NKX5
A0A2A4IZU8
A0A2H1W8P2
A0A194PU86
A0A2J7QT67
+ More
A0A067RDJ0 A0A1B0CJP6 A0A0K8TTZ7 A0A1A9WU46 A0A1I8PXH1 T1PAT1 A0A1A9VPG0 A0A1I8M7C7 A0A1B0AVK7 A0A2J7QT75 A0A1A9XU66 B4KCZ7 A0A146L3I8 B4LZI2 A0A1B0GCV6 A0A336LYK2 B4JUV3 B4GLI0 Q293X2 B3NZH2 A0A1W4ULW0 A0A3B0K5U1 B4HIC2 Q9VGR1 B4QU59 A0A1B6CLB2 B4PM32 A0A0L0BNZ5 A0A0A1WKT4 R4G5F2 U5EV90 A0A069DZE9 E9HAQ0 A0A0K8UC07 A0A034VN35 B4NIH2 A0A0K8T8F2 A0A0P5C1A5 A0A0A9YT43 A0A023F9E1 A0A1S3I5X6 A0A2J7QT60 W8BJG6 A0A2T7NXS5 V4AYZ5 A0A2J7QT41 A0A2J7QT65 A0A1B6GA87 A0A0A1WE29 A0A2C9KQA9 C3Y683 A0A2H8TT07 A0A1B6JTI9 E0VC45 Q3ZAL1 F4WIB3 A0A069DVV9 A0A195BLB6 A0A182VPT4 A0A1J1IG32 A0A224XLJ7 A0A182G1X6
A0A067RDJ0 A0A1B0CJP6 A0A0K8TTZ7 A0A1A9WU46 A0A1I8PXH1 T1PAT1 A0A1A9VPG0 A0A1I8M7C7 A0A1B0AVK7 A0A2J7QT75 A0A1A9XU66 B4KCZ7 A0A146L3I8 B4LZI2 A0A1B0GCV6 A0A336LYK2 B4JUV3 B4GLI0 Q293X2 B3NZH2 A0A1W4ULW0 A0A3B0K5U1 B4HIC2 Q9VGR1 B4QU59 A0A1B6CLB2 B4PM32 A0A0L0BNZ5 A0A0A1WKT4 R4G5F2 U5EV90 A0A069DZE9 E9HAQ0 A0A0K8UC07 A0A034VN35 B4NIH2 A0A0K8T8F2 A0A0P5C1A5 A0A0A9YT43 A0A023F9E1 A0A1S3I5X6 A0A2J7QT60 W8BJG6 A0A2T7NXS5 V4AYZ5 A0A2J7QT41 A0A2J7QT65 A0A1B6GA87 A0A0A1WE29 A0A2C9KQA9 C3Y683 A0A2H8TT07 A0A1B6JTI9 E0VC45 Q3ZAL1 F4WIB3 A0A069DVV9 A0A195BLB6 A0A182VPT4 A0A1J1IG32 A0A224XLJ7 A0A182G1X6
Pubmed
19121390
26354079
24845553
26369729
25315136
17994087
+ More
26823975 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 26108605 25830018 26334808 21292972 25348373 25401762 25474469 24495485 23254933 15562597 18563158 20566863 21719571 26483478
26823975 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 26108605 25830018 26334808 21292972 25348373 25401762 25474469 24495485 23254933 15562597 18563158 20566863 21719571 26483478
EMBL
BABH01030258
RSAL01000332
RVE42484.1
NWSH01004215
PCG65345.1
ODYU01006980
+ More
SOQ49316.1 KQ459592 KPI96882.1 NEVH01011202 PNF31763.1 KK852771 KDR16884.1 AJWK01014826 GDAI01000005 JAI17598.1 KA645028 AFP59657.1 JXJN01004287 PNF31764.1 CH933806 EDW13767.1 GDHC01015661 GDHC01007338 JAQ02968.1 JAQ11291.1 CH940650 EDW67121.1 CCAG010002237 UFQT01000290 SSX22880.1 CH916374 EDV91273.1 CH479185 EDW38404.1 CM000070 EAL29092.1 CH954181 EDV49545.1 OUUW01000007 SPP83420.1 CH480815 EDW42637.1 AE014297 AY118893 AAF54615.1 AAM50753.1 CM000364 EDX13390.1 GEDC01023041 JAS14257.1 CM000160 EDW96898.1 JRES01001582 KNC21731.1 GBXI01015162 JAC99129.1 GAHY01000021 JAA77489.1 GANO01003512 JAB56359.1 GBGD01000760 JAC88129.1 GL732612 EFX71220.1 GDHF01028112 JAI24202.1 GAKP01015063 JAC43889.1 CH964272 EDW84795.1 GBRD01003994 JAG61827.1 GDIP01177039 GDIQ01136140 JAJ46363.1 JAL15586.1 GBHO01010869 JAG32735.1 GBBI01001179 JAC17533.1 PNF31766.1 GAMC01005130 JAC01426.1 PZQS01000008 PVD25980.1 KB200129 ESP02933.1 PNF31761.1 PNF31767.1 GECZ01010427 JAS59342.1 GBXI01017614 JAC96677.1 GG666487 EEN64480.1 GFXV01005522 MBW17327.1 GECU01005237 JAT02470.1 DS235047 EEB10951.1 BT023858 AAZ86779.1 GL888175 EGI66044.1 GBGD01000844 JAC88045.1 KQ976440 KYM86480.1 CVRI01000047 CRK98492.1 GFTR01007403 JAW09023.1 JXUM01138109 JXUM01138110 KQ568682 KXJ68920.1
SOQ49316.1 KQ459592 KPI96882.1 NEVH01011202 PNF31763.1 KK852771 KDR16884.1 AJWK01014826 GDAI01000005 JAI17598.1 KA645028 AFP59657.1 JXJN01004287 PNF31764.1 CH933806 EDW13767.1 GDHC01015661 GDHC01007338 JAQ02968.1 JAQ11291.1 CH940650 EDW67121.1 CCAG010002237 UFQT01000290 SSX22880.1 CH916374 EDV91273.1 CH479185 EDW38404.1 CM000070 EAL29092.1 CH954181 EDV49545.1 OUUW01000007 SPP83420.1 CH480815 EDW42637.1 AE014297 AY118893 AAF54615.1 AAM50753.1 CM000364 EDX13390.1 GEDC01023041 JAS14257.1 CM000160 EDW96898.1 JRES01001582 KNC21731.1 GBXI01015162 JAC99129.1 GAHY01000021 JAA77489.1 GANO01003512 JAB56359.1 GBGD01000760 JAC88129.1 GL732612 EFX71220.1 GDHF01028112 JAI24202.1 GAKP01015063 JAC43889.1 CH964272 EDW84795.1 GBRD01003994 JAG61827.1 GDIP01177039 GDIQ01136140 JAJ46363.1 JAL15586.1 GBHO01010869 JAG32735.1 GBBI01001179 JAC17533.1 PNF31766.1 GAMC01005130 JAC01426.1 PZQS01000008 PVD25980.1 KB200129 ESP02933.1 PNF31761.1 PNF31767.1 GECZ01010427 JAS59342.1 GBXI01017614 JAC96677.1 GG666487 EEN64480.1 GFXV01005522 MBW17327.1 GECU01005237 JAT02470.1 DS235047 EEB10951.1 BT023858 AAZ86779.1 GL888175 EGI66044.1 GBGD01000844 JAC88045.1 KQ976440 KYM86480.1 CVRI01000047 CRK98492.1 GFTR01007403 JAW09023.1 JXUM01138109 JXUM01138110 KQ568682 KXJ68920.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000235965
UP000027135
+ More
UP000092461 UP000091820 UP000095300 UP000078200 UP000095301 UP000092460 UP000092443 UP000009192 UP000008792 UP000092444 UP000001070 UP000008744 UP000001819 UP000008711 UP000192221 UP000268350 UP000001292 UP000000803 UP000000304 UP000002282 UP000037069 UP000000305 UP000007798 UP000085678 UP000245119 UP000030746 UP000076420 UP000001554 UP000009046 UP000007755 UP000078540 UP000075920 UP000183832 UP000069940 UP000249989
UP000092461 UP000091820 UP000095300 UP000078200 UP000095301 UP000092460 UP000092443 UP000009192 UP000008792 UP000092444 UP000001070 UP000008744 UP000001819 UP000008711 UP000192221 UP000268350 UP000001292 UP000000803 UP000000304 UP000002282 UP000037069 UP000000305 UP000007798 UP000085678 UP000245119 UP000030746 UP000076420 UP000001554 UP000009046 UP000007755 UP000078540 UP000075920 UP000183832 UP000069940 UP000249989
Pfam
PF01465 GRIP
Interpro
IPR000237
GRIP_dom
ProteinModelPortal
H9J8P7
A0A3S2NKX5
A0A2A4IZU8
A0A2H1W8P2
A0A194PU86
A0A2J7QT67
+ More
A0A067RDJ0 A0A1B0CJP6 A0A0K8TTZ7 A0A1A9WU46 A0A1I8PXH1 T1PAT1 A0A1A9VPG0 A0A1I8M7C7 A0A1B0AVK7 A0A2J7QT75 A0A1A9XU66 B4KCZ7 A0A146L3I8 B4LZI2 A0A1B0GCV6 A0A336LYK2 B4JUV3 B4GLI0 Q293X2 B3NZH2 A0A1W4ULW0 A0A3B0K5U1 B4HIC2 Q9VGR1 B4QU59 A0A1B6CLB2 B4PM32 A0A0L0BNZ5 A0A0A1WKT4 R4G5F2 U5EV90 A0A069DZE9 E9HAQ0 A0A0K8UC07 A0A034VN35 B4NIH2 A0A0K8T8F2 A0A0P5C1A5 A0A0A9YT43 A0A023F9E1 A0A1S3I5X6 A0A2J7QT60 W8BJG6 A0A2T7NXS5 V4AYZ5 A0A2J7QT41 A0A2J7QT65 A0A1B6GA87 A0A0A1WE29 A0A2C9KQA9 C3Y683 A0A2H8TT07 A0A1B6JTI9 E0VC45 Q3ZAL1 F4WIB3 A0A069DVV9 A0A195BLB6 A0A182VPT4 A0A1J1IG32 A0A224XLJ7 A0A182G1X6
A0A067RDJ0 A0A1B0CJP6 A0A0K8TTZ7 A0A1A9WU46 A0A1I8PXH1 T1PAT1 A0A1A9VPG0 A0A1I8M7C7 A0A1B0AVK7 A0A2J7QT75 A0A1A9XU66 B4KCZ7 A0A146L3I8 B4LZI2 A0A1B0GCV6 A0A336LYK2 B4JUV3 B4GLI0 Q293X2 B3NZH2 A0A1W4ULW0 A0A3B0K5U1 B4HIC2 Q9VGR1 B4QU59 A0A1B6CLB2 B4PM32 A0A0L0BNZ5 A0A0A1WKT4 R4G5F2 U5EV90 A0A069DZE9 E9HAQ0 A0A0K8UC07 A0A034VN35 B4NIH2 A0A0K8T8F2 A0A0P5C1A5 A0A0A9YT43 A0A023F9E1 A0A1S3I5X6 A0A2J7QT60 W8BJG6 A0A2T7NXS5 V4AYZ5 A0A2J7QT41 A0A2J7QT65 A0A1B6GA87 A0A0A1WE29 A0A2C9KQA9 C3Y683 A0A2H8TT07 A0A1B6JTI9 E0VC45 Q3ZAL1 F4WIB3 A0A069DVV9 A0A195BLB6 A0A182VPT4 A0A1J1IG32 A0A224XLJ7 A0A182G1X6
Ontologies
GO
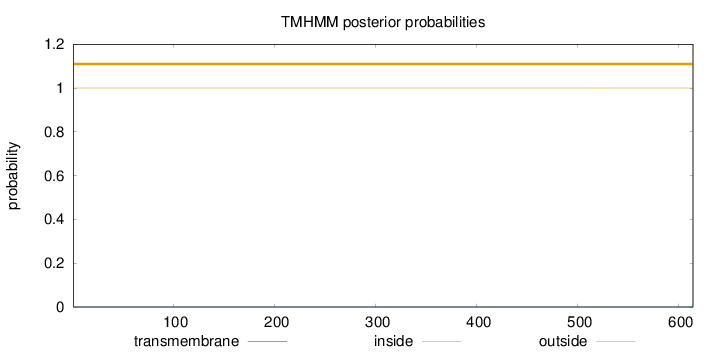
Topology
Length:
614
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00251
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00023
outside
1 - 614
Population Genetic Test Statistics
Pi
27.052178
Theta
24.626837
Tajima's D
0.241457
CLR
0.296323
CSRT
0.448477576121194
Interpretation
Uncertain
