Gene
KWMTBOMO07522
Pre Gene Modal
BGIBMGA005893
Annotation
PREDICTED:_CUGBP_Elav-like_family_member_4_isoform_X3_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.391 PlasmaMembrane Reliability : 1.244
Sequence
CDS
ATGCGAACGTTTAGCTGGCTCTGTATTACGCTCTCAGCTTTAGGTGCATCTTCAAGTCTGGTAGTGAAGTTCGCGGACACCGAGAAGGAACGACAGCTCCGACGTATGCAACAGATGGCAGGCAACATGGGCCTCTTGAATCCGTTCGTCTTCAACCAGTTCGGGACATACAACACTTACGCACAAGTCATCACTGAACAGCAACAAGCAGCGCTGATGGTGGCCGCGACGGCTCAAGGCTACATCAACCCTATGTCTGCGATCGCATCACACGCCCTCAACGGAATGACTAACTCTGTTGTACCGCCAACATCCGATAACTTTTCTGGTCTGGCGATAGGCACCGGTCAGCCTCTCAATGGTGCTCTACCATCTCTGCCGTCTCCTACTATGCCGGGCTTCAACATGGCTGCTCAGACTCCCAATGGACAACCACAGCCCCAAGAAGCGGTCTACACTAATGGCATTCACCAAACATTCACCGGACCCGTGCAGGTAACTCAAGGTCTACCCAACGGCGAGGCCGCAGCCCTGCAGCACGCGTACGGAGGGGGAATGCAGCCCTTCCCTGGCGTAGCTTACCCAGCGGTATATGGGCAGTTCCCACAGCCCATTCCACCACCGTTGTCCACCATTGCACCGGCCCAGCGAGAAGAATTTCTGATGTTCCCAGGCTGCTCCATTTCCGGCCCTGAGGGCTGTAACCTCTTCATTTACCACCTGCCCCAGGAATTTGGAGATGCCGAGCTCATGCAGATGTTTCTTCCCTTCGGCAACGTGATCAGCAGCAAAGTCTTTATCGATCGTGCCACTAACCAAAGTTTCGTGTCGTTCGATAACCCGGCGTCGGCGCAAGCTGCGATCCAAGCCATGAACGGCTTCCAGATCGGCATGAAGCGACTGAAGGTCCAGCTCAAACGGCCGAAGGACGCCAATCGGCCCTATTAA
Protein
MRTFSWLCITLSALGASSSLVVKFADTEKERQLRRMQQMAGNMGLLNPFVFNQFGTYNTYAQVITEQQQAALMVAATAQGYINPMSAIASHALNGMTNSVVPPTSDNFSGLAIGTGQPLNGALPSLPSPTMPGFNMAAQTPNGQPQPQEAVYTNGIHQTFTGPVQVTQGLPNGEAAALQHAYGGGMQPFPGVAYPAVYGQFPQPIPPPLSTIAPAQREEFLMFPGCSISGPEGCNLFIYHLPQEFGDAELMQMFLPFGNVISSKVFIDRATNQSFVSFDNPASAQAAIQAMNGFQIGMKRLKVQLKRPKDANRPY
Summary
Uniprot
A0A2A4J7Z9
A0A3S2M2C3
A0A2H1VFP5
A0A2A4J721
H9J8Q1
A0A194PV61
+ More
A0A212FBK0 A0A139WAW6 A0A139WB31 A0A0N1PGB3 A0A139WAW4 A0A1Y1L3S4 A0A1B6CN79 A0A1B6CU29 A0A1B6CX00 A0A1B6CYQ9 A0A0M9AAP6 A0A1B6DK86 A0A1B6E8C2 A0A2C9GQ67 A0A1B6DKL5 A0A1B6DW12 U5EPM1 A0A1B6E1N2 A0A1B6C5D8 A0A3L8DKY3 A0A1B6D8E4 A0A1B6C710 A0A0L7R4D9 V9IDC0 A0A088AVG9 A0A1S4GML9 Q7PPI2 A0A2A3EJS5 Q17E15 Q16HG5 A0A084WE23 A0A336MCH9 A0A0K8V2P4 A0A1W4VIU8 A0A0J9RV49 A0A0R1E2H8 A0A0Q5U4K4 A0A154P4I7 A0A1W4V738 A0A1W4VIT8 A0A0J9UKM3 A0A0Q5UIV4 A0A1W4VJF3 A0A0J9RW52 A0A0R1E2N5 A0A0Q5UB24 A0A1W4VK77 A0A0R1E2P1 A0A0Q5UJ32 A0A0Q9WTM2 A0A0J9RUV1 A0A0R1DXS7 A0A0Q5UJB6 A0A0Q9XBY1 A0A0J9RUU3 A0A0R1DXM1 A0A0Q5U4T6 A0A0Q9XL53 A0A0Q9WJB5 A0A0Q9XNT7 A0A0Q9WJS1 A0A0Q9XBX6 A0A0J9RUU7 M9MS36 A0A0R1DXH9 A0A0Q5U6V6 A0A0J9RW45 A0A1W4WMP7 A0A0J9RV36 A0A0Q9XM96 A0A0Q9WVI4 A0A0Q9XNR2 A0A0Q9WVM0 A0A0Q9XL39 B4LGT9 B4L124 B4IXL1 A0A0Q9XC25 A0A158NMS5 W5JV15 A0A158NMS4 A0A1W4V6Q1 A0A0P9AMP9 A0A0N8P163 A0A1W4VJG6 A0A0P9C5V7 A0A0P8YJA3 A0A1W4VK91 A0A0P8XWE5 A0A1W4VK82 A0A0P8XWH6 A0A1W4V6N6 B3MAP2 A0A0J9RUM3
A0A212FBK0 A0A139WAW6 A0A139WB31 A0A0N1PGB3 A0A139WAW4 A0A1Y1L3S4 A0A1B6CN79 A0A1B6CU29 A0A1B6CX00 A0A1B6CYQ9 A0A0M9AAP6 A0A1B6DK86 A0A1B6E8C2 A0A2C9GQ67 A0A1B6DKL5 A0A1B6DW12 U5EPM1 A0A1B6E1N2 A0A1B6C5D8 A0A3L8DKY3 A0A1B6D8E4 A0A1B6C710 A0A0L7R4D9 V9IDC0 A0A088AVG9 A0A1S4GML9 Q7PPI2 A0A2A3EJS5 Q17E15 Q16HG5 A0A084WE23 A0A336MCH9 A0A0K8V2P4 A0A1W4VIU8 A0A0J9RV49 A0A0R1E2H8 A0A0Q5U4K4 A0A154P4I7 A0A1W4V738 A0A1W4VIT8 A0A0J9UKM3 A0A0Q5UIV4 A0A1W4VJF3 A0A0J9RW52 A0A0R1E2N5 A0A0Q5UB24 A0A1W4VK77 A0A0R1E2P1 A0A0Q5UJ32 A0A0Q9WTM2 A0A0J9RUV1 A0A0R1DXS7 A0A0Q5UJB6 A0A0Q9XBY1 A0A0J9RUU3 A0A0R1DXM1 A0A0Q5U4T6 A0A0Q9XL53 A0A0Q9WJB5 A0A0Q9XNT7 A0A0Q9WJS1 A0A0Q9XBX6 A0A0J9RUU7 M9MS36 A0A0R1DXH9 A0A0Q5U6V6 A0A0J9RW45 A0A1W4WMP7 A0A0J9RV36 A0A0Q9XM96 A0A0Q9WVI4 A0A0Q9XNR2 A0A0Q9WVM0 A0A0Q9XL39 B4LGT9 B4L124 B4IXL1 A0A0Q9XC25 A0A158NMS5 W5JV15 A0A158NMS4 A0A1W4V6Q1 A0A0P9AMP9 A0A0N8P163 A0A1W4VJG6 A0A0P9C5V7 A0A0P8YJA3 A0A1W4VK91 A0A0P8XWE5 A0A1W4VK82 A0A0P8XWH6 A0A1W4V6N6 B3MAP2 A0A0J9RUM3
Pubmed
EMBL
NWSH01002597
PCG67886.1
RSAL01000060
RVE49635.1
ODYU01002114
SOQ39202.1
+ More
PCG67885.1 BABH01030357 KQ459592 KPI96878.1 AGBW02009309 OWR51109.1 KQ971379 KYB25052.1 KYB25051.1 KQ460954 KPJ10261.1 KYB25053.1 GEZM01067693 JAV67441.1 GEDC01022503 JAS14795.1 GEDC01020292 JAS17006.1 GEDC01019575 JAS17723.1 GEDC01018905 JAS18393.1 KQ435698 KOX80657.1 GEDC01011233 JAS26065.1 GEDC01003125 JAS34173.1 APCN01004645 APCN01004646 GEDC01011138 JAS26160.1 GEDC01007448 JAS29850.1 GANO01003665 JAB56206.1 GEDC01005501 JAS31797.1 GEDC01028684 JAS08614.1 QOIP01000007 RLU21077.1 GEDC01015378 JAS21920.1 GEDC01028179 JAS09119.1 KQ414657 KOC65750.1 JR039326 AEY58642.1 AAAB01008948 EAA10371.5 KZ288240 PBC31291.1 CH477288 EAT44684.1 CH478174 EAT33691.1 ATLV01023117 ATLV01023118 KE525340 KFB48467.1 UFQS01000763 UFQT01000763 SSX06705.1 SSX27051.1 GDHF01019130 JAI33184.1 CM002912 KMY99457.1 CM000159 KRK01826.1 CH954178 KQS43909.1 KQ434810 KZC06742.1 KMY99455.1 KQS43910.1 KMY99454.1 KRK01818.1 KQS43901.1 KRK01828.1 KQS43914.1 CH940647 KRF84977.1 KRF84984.1 KMY99458.1 KRK01817.1 KQS43904.1 KQS43919.1 CH933809 KRG06054.1 KMY99448.1 KRK01820.1 KRK01829.1 KQS43899.1 KRG06052.1 KRF84988.1 KRG06055.1 KRF84985.1 KRG06027.1 KMY99453.1 AE014296 ADV37531.1 KRK01814.1 KQS43900.1 KMY99440.1 KMY99444.1 KMY99442.1 KMY99450.1 KRG06030.1 KRF84980.1 KRG06023.1 KRF84990.1 KRG06036.1 EDW70554.1 EDW18181.2 CH916366 EDV97473.1 KRG06028.1 ADTU01020719 ADTU01020720 ADTU01020721 ADTU01020722 ADTU01020723 ADTU01020724 ADMH02000375 ETN66795.1 CH902618 KPU79051.1 KPU79044.1 KPU79047.1 KPU79055.1 KPU79043.1 KPU79050.1 EDV41329.1 KPU79041.1 KMY99461.1
PCG67885.1 BABH01030357 KQ459592 KPI96878.1 AGBW02009309 OWR51109.1 KQ971379 KYB25052.1 KYB25051.1 KQ460954 KPJ10261.1 KYB25053.1 GEZM01067693 JAV67441.1 GEDC01022503 JAS14795.1 GEDC01020292 JAS17006.1 GEDC01019575 JAS17723.1 GEDC01018905 JAS18393.1 KQ435698 KOX80657.1 GEDC01011233 JAS26065.1 GEDC01003125 JAS34173.1 APCN01004645 APCN01004646 GEDC01011138 JAS26160.1 GEDC01007448 JAS29850.1 GANO01003665 JAB56206.1 GEDC01005501 JAS31797.1 GEDC01028684 JAS08614.1 QOIP01000007 RLU21077.1 GEDC01015378 JAS21920.1 GEDC01028179 JAS09119.1 KQ414657 KOC65750.1 JR039326 AEY58642.1 AAAB01008948 EAA10371.5 KZ288240 PBC31291.1 CH477288 EAT44684.1 CH478174 EAT33691.1 ATLV01023117 ATLV01023118 KE525340 KFB48467.1 UFQS01000763 UFQT01000763 SSX06705.1 SSX27051.1 GDHF01019130 JAI33184.1 CM002912 KMY99457.1 CM000159 KRK01826.1 CH954178 KQS43909.1 KQ434810 KZC06742.1 KMY99455.1 KQS43910.1 KMY99454.1 KRK01818.1 KQS43901.1 KRK01828.1 KQS43914.1 CH940647 KRF84977.1 KRF84984.1 KMY99458.1 KRK01817.1 KQS43904.1 KQS43919.1 CH933809 KRG06054.1 KMY99448.1 KRK01820.1 KRK01829.1 KQS43899.1 KRG06052.1 KRF84988.1 KRG06055.1 KRF84985.1 KRG06027.1 KMY99453.1 AE014296 ADV37531.1 KRK01814.1 KQS43900.1 KMY99440.1 KMY99444.1 KMY99442.1 KMY99450.1 KRG06030.1 KRF84980.1 KRG06023.1 KRF84990.1 KRG06036.1 EDW70554.1 EDW18181.2 CH916366 EDV97473.1 KRG06028.1 ADTU01020719 ADTU01020720 ADTU01020721 ADTU01020722 ADTU01020723 ADTU01020724 ADMH02000375 ETN66795.1 CH902618 KPU79051.1 KPU79044.1 KPU79047.1 KPU79055.1 KPU79043.1 KPU79050.1 EDV41329.1 KPU79041.1 KMY99461.1
Proteomes
UP000218220
UP000283053
UP000005204
UP000053268
UP000007151
UP000007266
+ More
UP000053240 UP000053105 UP000075840 UP000279307 UP000053825 UP000005203 UP000007062 UP000242457 UP000008820 UP000030765 UP000192221 UP000002282 UP000008711 UP000076502 UP000008792 UP000009192 UP000000803 UP000192223 UP000001070 UP000005205 UP000000673 UP000007801
UP000053240 UP000053105 UP000075840 UP000279307 UP000053825 UP000005203 UP000007062 UP000242457 UP000008820 UP000030765 UP000192221 UP000002282 UP000008711 UP000076502 UP000008792 UP000009192 UP000000803 UP000192223 UP000001070 UP000005205 UP000000673 UP000007801
PRIDE
Pfam
PF00076 RRM_1
SUPFAM
SSF54928
SSF54928
Gene 3D
ProteinModelPortal
A0A2A4J7Z9
A0A3S2M2C3
A0A2H1VFP5
A0A2A4J721
H9J8Q1
A0A194PV61
+ More
A0A212FBK0 A0A139WAW6 A0A139WB31 A0A0N1PGB3 A0A139WAW4 A0A1Y1L3S4 A0A1B6CN79 A0A1B6CU29 A0A1B6CX00 A0A1B6CYQ9 A0A0M9AAP6 A0A1B6DK86 A0A1B6E8C2 A0A2C9GQ67 A0A1B6DKL5 A0A1B6DW12 U5EPM1 A0A1B6E1N2 A0A1B6C5D8 A0A3L8DKY3 A0A1B6D8E4 A0A1B6C710 A0A0L7R4D9 V9IDC0 A0A088AVG9 A0A1S4GML9 Q7PPI2 A0A2A3EJS5 Q17E15 Q16HG5 A0A084WE23 A0A336MCH9 A0A0K8V2P4 A0A1W4VIU8 A0A0J9RV49 A0A0R1E2H8 A0A0Q5U4K4 A0A154P4I7 A0A1W4V738 A0A1W4VIT8 A0A0J9UKM3 A0A0Q5UIV4 A0A1W4VJF3 A0A0J9RW52 A0A0R1E2N5 A0A0Q5UB24 A0A1W4VK77 A0A0R1E2P1 A0A0Q5UJ32 A0A0Q9WTM2 A0A0J9RUV1 A0A0R1DXS7 A0A0Q5UJB6 A0A0Q9XBY1 A0A0J9RUU3 A0A0R1DXM1 A0A0Q5U4T6 A0A0Q9XL53 A0A0Q9WJB5 A0A0Q9XNT7 A0A0Q9WJS1 A0A0Q9XBX6 A0A0J9RUU7 M9MS36 A0A0R1DXH9 A0A0Q5U6V6 A0A0J9RW45 A0A1W4WMP7 A0A0J9RV36 A0A0Q9XM96 A0A0Q9WVI4 A0A0Q9XNR2 A0A0Q9WVM0 A0A0Q9XL39 B4LGT9 B4L124 B4IXL1 A0A0Q9XC25 A0A158NMS5 W5JV15 A0A158NMS4 A0A1W4V6Q1 A0A0P9AMP9 A0A0N8P163 A0A1W4VJG6 A0A0P9C5V7 A0A0P8YJA3 A0A1W4VK91 A0A0P8XWE5 A0A1W4VK82 A0A0P8XWH6 A0A1W4V6N6 B3MAP2 A0A0J9RUM3
A0A212FBK0 A0A139WAW6 A0A139WB31 A0A0N1PGB3 A0A139WAW4 A0A1Y1L3S4 A0A1B6CN79 A0A1B6CU29 A0A1B6CX00 A0A1B6CYQ9 A0A0M9AAP6 A0A1B6DK86 A0A1B6E8C2 A0A2C9GQ67 A0A1B6DKL5 A0A1B6DW12 U5EPM1 A0A1B6E1N2 A0A1B6C5D8 A0A3L8DKY3 A0A1B6D8E4 A0A1B6C710 A0A0L7R4D9 V9IDC0 A0A088AVG9 A0A1S4GML9 Q7PPI2 A0A2A3EJS5 Q17E15 Q16HG5 A0A084WE23 A0A336MCH9 A0A0K8V2P4 A0A1W4VIU8 A0A0J9RV49 A0A0R1E2H8 A0A0Q5U4K4 A0A154P4I7 A0A1W4V738 A0A1W4VIT8 A0A0J9UKM3 A0A0Q5UIV4 A0A1W4VJF3 A0A0J9RW52 A0A0R1E2N5 A0A0Q5UB24 A0A1W4VK77 A0A0R1E2P1 A0A0Q5UJ32 A0A0Q9WTM2 A0A0J9RUV1 A0A0R1DXS7 A0A0Q5UJB6 A0A0Q9XBY1 A0A0J9RUU3 A0A0R1DXM1 A0A0Q5U4T6 A0A0Q9XL53 A0A0Q9WJB5 A0A0Q9XNT7 A0A0Q9WJS1 A0A0Q9XBX6 A0A0J9RUU7 M9MS36 A0A0R1DXH9 A0A0Q5U6V6 A0A0J9RW45 A0A1W4WMP7 A0A0J9RV36 A0A0Q9XM96 A0A0Q9WVI4 A0A0Q9XNR2 A0A0Q9WVM0 A0A0Q9XL39 B4LGT9 B4L124 B4IXL1 A0A0Q9XC25 A0A158NMS5 W5JV15 A0A158NMS4 A0A1W4V6Q1 A0A0P9AMP9 A0A0N8P163 A0A1W4VJG6 A0A0P9C5V7 A0A0P8YJA3 A0A1W4VK91 A0A0P8XWE5 A0A1W4VK82 A0A0P8XWH6 A0A1W4V6N6 B3MAP2 A0A0J9RUM3
PDB
4TLQ
E-value=1.72669e-32,
Score=347
Ontologies
GO
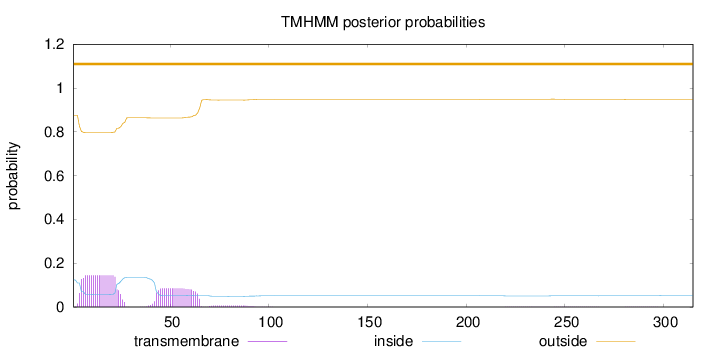
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.960236
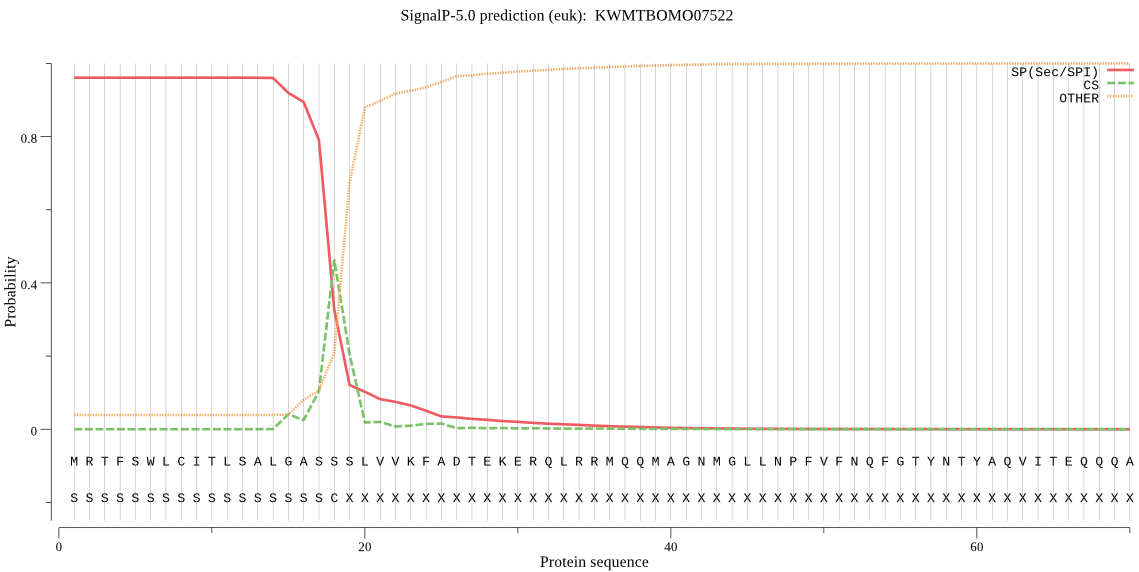
Length:
315
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.9727
Exp number, first 60 AAs:
4.47983
Total prob of N-in:
0.12477
outside
1 - 315
Population Genetic Test Statistics
Pi
13.938833
Theta
17.428982
Tajima's D
-0.792958
CLR
1.943985
CSRT
0.181040947952602
Interpretation
Uncertain
