Gene
KWMTBOMO07521
Pre Gene Modal
BGIBMGA005885
Annotation
PREDICTED:_poly(ADP-ribose)_glycohydrolase_[Bombyx_mori]
Full name
Poly(ADP-ribose) glycohydrolase
Location in the cell
Nuclear Reliability : 3.504
Sequence
CDS
ATGAGTACAGTAACTAAAAGCTGGCTCGGAGTACCCATTAGCGAAATACGCGGTTCCCAATCGCCGTGGGGTGCTCCAGAATTTCCGTTAGTGGTGCCAGCATACAATCATTCCGTTTTGTATCACGTGCAGAACGGTAGACCGATTGATGGAGACAGACCGCCAAAACCGCAGATAGGACGTGATAAGTGGGATCATGAGTTTGTAAGGATGCCGTGTTCAAATCAGAGCCTTTATCCATTTGAAGATAGCAACGGTGATTGCCACTTGAAAAAGAGATGGGAGGTTATAGAGAGTAGTCTCTGGAAACCTATCAGGGACAGTCGGGAACTGGCTAACACTATACTCAGCTACAATACAAAGTTTAAGAAAATATGGAAATTCAAAGCCTTACACAAATTGTTCAGTGAATATCTAGATGAAGAAGAAACACAATACTTCTTTGATGTTACATTACCTGAGATAGCGAAGTTAGCGTTGGCATTGCCAAAATTGATTCAGTCACCCATACCACTACTGAAACAACACAAGAACTATTCTATATCGTTGTCACAACAACAAATAGCAAGTATCCTTGCGAATGCTTTTTTCTGTACTTTTCCACGACGGAATACAAAAAAGAAATCATCTGAATACTCATCTTATCCACACATCAATTTTTATACATTGTACGAGACAGATGGTTCTGAATCAGTGTTAGAAAAGTTAAAATGTTTGTGTCACTACTTTCGAAGAGTGTGCACTAAAGTCCCGGGCGGCGTAGTGACTTGGACCCGGCGCAGCGTGCCGCCGCGGGGCTGCCCCGCGTGGGGCTCCAGTGGGTCTCCGCTGTCGTCCCTCCCACTGCACGTAGACTCCGACACCACTATAGAGGATGCTCACGGACTCATACAGGTCGACTTCGCTAACAAGTACTTAGGTGGTGGTGTATTGAACTATGGTTGCGTGCAAGAAGAGATAAGATTCGTGATATGTCCAGAGCTGATGGTGTCGATGCTGTTCACGGAAGTGCTGAGGCCCAATGAAGCATTGTTAATTGTTGGCTGTGAAAGGTACAGCAGGTACACGGGCTACGCGAGCACGTTCCAGTGGGCCGGGGACTACCGGGACGAGACGCCCCTGGACTCGTCCGGGCGGCGGCGCTGCGCCGTGCTGGCCATAGACGCGCTGCCCTACCCCAGCACCGTCCAGGAGTACAGGGAGGAAATGTTGACCAGGGAACTTAATAAGGCCTGGATAGGTTTTACGTTTCACGCGGACCCAGAGTCCAGTACGCTGCAGTACCCGGGCGTGGCGACCGGCAACTGGGGCTGCGGCGCCTTCGGGGGCTCCCCGCGGCTCAAGGGTCTCCTGCAGGCGATGGCGGCCGCGCAGGCCGGGCGGCCCCTCGCTTACTTTACGTTCGGCGACGCGGAGCTCAGGGACCACATCGTAGACGTGTACAACGCGCTGGCCAAGTGTGGCGTCACCGTCGGTCAGCTGTACCGACTCATTGTTCGTTTCTCAAAAACGAATGTACTCAGGTCGGACCTTTACGATTTTTTGCATCAGGTGTTGAGAGACGGGGAGGTGGATAAGCTCAGGGGAACTCAGCACGAGCGCATGGAGACCGACTCAACACCGGGCACGTCGACAATGGAACTGTCCGACTCCCTACAGACGGAATTACTCAAAGAGTTTACATCGACCGATTCACCGGACCTATTCTCGACTGACGAAAGTTTAATCGAGATTAAAATCGATAAGAGCGATCCGAGTGACAACACTGGCTGTCACTCGCAGCTGTCAAACTGTACGTCAAAGTTGTTTGACGAGATGCAGAAGCTGGATGAGGAAGACGGGCTGACGAGCTTGTCTATGCCCCAAGAGTCGCCTCTGGAACAGAACGCGAGTTGCGACGGCGATCTGCCCAAGCGACCCGCAGATGCGACTTCGGAATTGAAGGCCAGAGTCAATAGGAAAATAACAGATTATTTTAGCAGAAAACCTGTTTAA
Protein
MSTVTKSWLGVPISEIRGSQSPWGAPEFPLVVPAYNHSVLYHVQNGRPIDGDRPPKPQIGRDKWDHEFVRMPCSNQSLYPFEDSNGDCHLKKRWEVIESSLWKPIRDSRELANTILSYNTKFKKIWKFKALHKLFSEYLDEEETQYFFDVTLPEIAKLALALPKLIQSPIPLLKQHKNYSISLSQQQIASILANAFFCTFPRRNTKKKSSEYSSYPHINFYTLYETDGSESVLEKLKCLCHYFRRVCTKVPGGVVTWTRRSVPPRGCPAWGSSGSPLSSLPLHVDSDTTIEDAHGLIQVDFANKYLGGGVLNYGCVQEEIRFVICPELMVSMLFTEVLRPNEALLIVGCERYSRYTGYASTFQWAGDYRDETPLDSSGRRRCAVLAIDALPYPSTVQEYREEMLTRELNKAWIGFTFHADPESSTLQYPGVATGNWGCGAFGGSPRLKGLLQAMAAAQAGRPLAYFTFGDAELRDHIVDVYNALAKCGVTVGQLYRLIVRFSKTNVLRSDLYDFLHQVLRDGEVDKLRGTQHERMETDSTPGTSTMELSDSLQTELLKEFTSTDSPDLFSTDESLIEIKIDKSDPSDNTGCHSQLSNCTSKLFDEMQKLDEEDGLTSLSMPQESPLEQNASCDGDLPKRPADATSELKARVNRKITDYFSRKPV
Summary
Description
Poly(ADP-ribose) synthesized after DNA damage is only present transiently and is rapidly degraded by poly(ADP-ribose) glycohydrolase. Poly(ADP-ribose) metabolism is required for maintenance of the normal function of neuronal cells.
Catalytic Activity
[(1''->2')-ADP-alpha-D-ribose]n + H2O = [(1''->2')-ADP-alpha-D-ribose]n-1 + ADP-D-ribose
Similarity
Belongs to the poly(ADP-ribose) glycohydrolase family.
Keywords
Complete proteome
Hydrolase
Phosphoprotein
Reference proteome
Feature
chain Poly(ADP-ribose) glycohydrolase
Uniprot
H9J8P3
A0A3S2NV82
A0A2A4J6Y6
A0A194PU76
A0A212ER22
B0WST7
+ More
A0A1Q3F4V3 A0A1I8P7Y8 Q17L69 A0A0L0C4D8 A0A0A1WZR8 A0A182GTV7 B4R467 A0A212FBJ9 A0A034W105 O46043 A0A1A9W8L6 A0A1W4VSH3 A0A0K8UU75 A0A1B0ADR4 A0A1A9VY97 A0A0K8UFZ1 W8AQL3 A0A1B0FJ80 B3NSQ9 A0A1I8NDE2 B4Q0I7 A0A182GBX4 A0A1I8NDE8 U5EQC3 B4NPN3 B4I158 A0A1L8DL32 B4M1L2 A0A3B0JWQ9 B4H0U9 A0A3B0JRR4 Q29GN0 A0A1B0GHX2 B3MW84 A0A0M4F7R8 K7J0P1 B4JN37 B4L4V6 A0A0C9QWZ8 A0A232F6A4 A0A3L8DI25 A0A2J7PYB3 A0A2J7PY66 E2BI70 A0A1B0DM96 A0A2J7PY95 A0A158NCD2 A0A088AIJ7 F4WWX1 A0A1B6KY88 A0A2J7PY76 A0A336MPT7 A0A1B6HT42 A0A026VX02 A0A1B6HAF6 A0A1J1HID5 D6W990 E2B1V8 A0A067RFC3 A0A2P8Z7V8 A0A2A3EK29 A0A1W4WD06 A0A0P5Y1S9 A0A0P5Q659 A0A195FAJ8 A0A0P5NMJ0 A0A195BY71 A0A151WS21 A0A151ITL5 A0A0N8BQQ5 A0A0P5QQE3 A0A069DVA2 A0A0P6K040 A0A2S2QZC1 E9GAG4 A0A2G8K0Z3 A0A151IG65 A0A1Y1KZH8 A0A154PFY3 A0A2D4MHK1 H9GUD9 A0A210PSB1 A0A3Q7RU58 J9NTQ1 A0A2U3X9T8 A0A2U3X9S9 A0A2Y9HZM2 A0A340WI96 A0A3Q7PSD1 A0A340WBF8 A0A0F7ZDZ9 K9IUD8
A0A1Q3F4V3 A0A1I8P7Y8 Q17L69 A0A0L0C4D8 A0A0A1WZR8 A0A182GTV7 B4R467 A0A212FBJ9 A0A034W105 O46043 A0A1A9W8L6 A0A1W4VSH3 A0A0K8UU75 A0A1B0ADR4 A0A1A9VY97 A0A0K8UFZ1 W8AQL3 A0A1B0FJ80 B3NSQ9 A0A1I8NDE2 B4Q0I7 A0A182GBX4 A0A1I8NDE8 U5EQC3 B4NPN3 B4I158 A0A1L8DL32 B4M1L2 A0A3B0JWQ9 B4H0U9 A0A3B0JRR4 Q29GN0 A0A1B0GHX2 B3MW84 A0A0M4F7R8 K7J0P1 B4JN37 B4L4V6 A0A0C9QWZ8 A0A232F6A4 A0A3L8DI25 A0A2J7PYB3 A0A2J7PY66 E2BI70 A0A1B0DM96 A0A2J7PY95 A0A158NCD2 A0A088AIJ7 F4WWX1 A0A1B6KY88 A0A2J7PY76 A0A336MPT7 A0A1B6HT42 A0A026VX02 A0A1B6HAF6 A0A1J1HID5 D6W990 E2B1V8 A0A067RFC3 A0A2P8Z7V8 A0A2A3EK29 A0A1W4WD06 A0A0P5Y1S9 A0A0P5Q659 A0A195FAJ8 A0A0P5NMJ0 A0A195BY71 A0A151WS21 A0A151ITL5 A0A0N8BQQ5 A0A0P5QQE3 A0A069DVA2 A0A0P6K040 A0A2S2QZC1 E9GAG4 A0A2G8K0Z3 A0A151IG65 A0A1Y1KZH8 A0A154PFY3 A0A2D4MHK1 H9GUD9 A0A210PSB1 A0A3Q7RU58 J9NTQ1 A0A2U3X9T8 A0A2U3X9S9 A0A2Y9HZM2 A0A340WI96 A0A3Q7PSD1 A0A340WBF8 A0A0F7ZDZ9 K9IUD8
EC Number
3.2.1.143
Pubmed
19121390
26354079
22118469
17510324
26108605
25830018
+ More
26483478 17994087 25348373 10731132 12537572 10731137 12537569 14676324 18327897 30100084 24495485 25315136 17550304 15632085 20075255 28648823 30249741 20798317 21347285 21719571 24508170 18362917 19820115 24845553 29403074 26334808 21292972 29023486 28004739 21881562 28812685 16341006
26483478 17994087 25348373 10731132 12537572 10731137 12537569 14676324 18327897 30100084 24495485 25315136 17550304 15632085 20075255 28648823 30249741 20798317 21347285 21719571 24508170 18362917 19820115 24845553 29403074 26334808 21292972 29023486 28004739 21881562 28812685 16341006
EMBL
BABH01030360
BABH01030361
RSAL01000060
RVE49636.1
NWSH01002597
PCG67887.1
+ More
KQ459592 KPI96872.1 AGBW02013163 OWR43927.1 DS232076 EDS34047.1 GFDL01012478 JAV22567.1 CH477218 EAT47426.1 JRES01000996 KNC26314.1 GBXI01009723 JAD04569.1 JXUM01087914 KQ563665 KXJ73521.1 CM000366 EDX17009.1 AGBW02009309 OWR51110.1 GAKP01010925 JAC48027.1 AF079556 AE014298 Z98254 AY051955 AAC28734.1 AAK93379.1 CAB10913.1 GDHF01022090 JAI30224.1 GDHF01026831 JAI25483.1 GAMC01015445 JAB91110.1 CCAG010011096 CH954180 EDV45739.2 CM000162 EDX01271.2 JXUM01053377 KQ561774 KXJ77584.1 GANO01003331 JAB56540.1 CH964291 EDW86473.2 CH480819 EDW53239.1 GFDF01006915 JAV07169.1 CH940651 EDW65566.2 OUUW01000003 SPP78169.1 CH479201 EDW30014.1 SPP78170.1 CH379064 EAL32079.3 AJWK01006336 CH902625 EDV35229.1 CP012528 ALC48287.1 CH916371 EDV92130.1 CH933810 EDW07584.2 GBYB01000215 JAG69982.1 NNAY01000903 OXU26003.1 QOIP01000008 RLU19976.1 NEVH01020850 PNF21290.1 PNF21289.1 GL448457 EFN84601.1 AJVK01006914 PNF21292.1 ADTU01011818 GL888417 EGI61214.1 GEBQ01023753 JAT16224.1 PNF21291.1 UFQS01001383 UFQT01001383 SSX10624.1 SSX30307.1 GECU01029885 JAS77821.1 KK107648 EZA48323.1 GECU01036079 JAS71627.1 CVRI01000004 CRK87314.1 KQ971312 EEZ98465.1 GL444981 EFN60314.1 KK852722 KDR17705.1 PYGN01000157 PSN52583.1 KZ288222 PBC32050.1 GDIP01065195 LRGB01000389 JAM38520.1 KZS19345.1 GDIQ01121524 JAL30202.1 KQ981727 KYN37069.1 GDIQ01147230 JAL04496.1 KQ976396 KYM92888.1 KQ982796 KYQ50618.1 KQ981004 KYN10390.1 GDIQ01154219 JAK97506.1 GDIQ01117442 JAL34284.1 GBGD01000886 JAC88003.1 GDIQ01001081 JAN93656.1 GGMS01013896 MBY83099.1 GL732537 EFX83529.1 MRZV01001001 PIK41639.1 KQ977728 KYN00259.1 GEZM01068670 GEZM01068669 JAV66813.1 KQ434896 KZC10773.1 IACM01100102 LAB32246.1 AAWZ02012964 AAWZ02012965 NEDP02005529 OWF39385.1 AAEX03015370 GBEX01000128 JAI14432.1 GABZ01000620 JAA52905.1
KQ459592 KPI96872.1 AGBW02013163 OWR43927.1 DS232076 EDS34047.1 GFDL01012478 JAV22567.1 CH477218 EAT47426.1 JRES01000996 KNC26314.1 GBXI01009723 JAD04569.1 JXUM01087914 KQ563665 KXJ73521.1 CM000366 EDX17009.1 AGBW02009309 OWR51110.1 GAKP01010925 JAC48027.1 AF079556 AE014298 Z98254 AY051955 AAC28734.1 AAK93379.1 CAB10913.1 GDHF01022090 JAI30224.1 GDHF01026831 JAI25483.1 GAMC01015445 JAB91110.1 CCAG010011096 CH954180 EDV45739.2 CM000162 EDX01271.2 JXUM01053377 KQ561774 KXJ77584.1 GANO01003331 JAB56540.1 CH964291 EDW86473.2 CH480819 EDW53239.1 GFDF01006915 JAV07169.1 CH940651 EDW65566.2 OUUW01000003 SPP78169.1 CH479201 EDW30014.1 SPP78170.1 CH379064 EAL32079.3 AJWK01006336 CH902625 EDV35229.1 CP012528 ALC48287.1 CH916371 EDV92130.1 CH933810 EDW07584.2 GBYB01000215 JAG69982.1 NNAY01000903 OXU26003.1 QOIP01000008 RLU19976.1 NEVH01020850 PNF21290.1 PNF21289.1 GL448457 EFN84601.1 AJVK01006914 PNF21292.1 ADTU01011818 GL888417 EGI61214.1 GEBQ01023753 JAT16224.1 PNF21291.1 UFQS01001383 UFQT01001383 SSX10624.1 SSX30307.1 GECU01029885 JAS77821.1 KK107648 EZA48323.1 GECU01036079 JAS71627.1 CVRI01000004 CRK87314.1 KQ971312 EEZ98465.1 GL444981 EFN60314.1 KK852722 KDR17705.1 PYGN01000157 PSN52583.1 KZ288222 PBC32050.1 GDIP01065195 LRGB01000389 JAM38520.1 KZS19345.1 GDIQ01121524 JAL30202.1 KQ981727 KYN37069.1 GDIQ01147230 JAL04496.1 KQ976396 KYM92888.1 KQ982796 KYQ50618.1 KQ981004 KYN10390.1 GDIQ01154219 JAK97506.1 GDIQ01117442 JAL34284.1 GBGD01000886 JAC88003.1 GDIQ01001081 JAN93656.1 GGMS01013896 MBY83099.1 GL732537 EFX83529.1 MRZV01001001 PIK41639.1 KQ977728 KYN00259.1 GEZM01068670 GEZM01068669 JAV66813.1 KQ434896 KZC10773.1 IACM01100102 LAB32246.1 AAWZ02012964 AAWZ02012965 NEDP02005529 OWF39385.1 AAEX03015370 GBEX01000128 JAI14432.1 GABZ01000620 JAA52905.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000007151
UP000002320
+ More
UP000095300 UP000008820 UP000037069 UP000069940 UP000249989 UP000000304 UP000000803 UP000091820 UP000192221 UP000092445 UP000078200 UP000092444 UP000008711 UP000095301 UP000002282 UP000007798 UP000001292 UP000008792 UP000268350 UP000008744 UP000001819 UP000092461 UP000007801 UP000092553 UP000002358 UP000001070 UP000009192 UP000215335 UP000279307 UP000235965 UP000008237 UP000092462 UP000005205 UP000005203 UP000007755 UP000053097 UP000183832 UP000007266 UP000000311 UP000027135 UP000245037 UP000242457 UP000192223 UP000076858 UP000078541 UP000078540 UP000075809 UP000078492 UP000000305 UP000230750 UP000078542 UP000076502 UP000001646 UP000242188 UP000286640 UP000002254 UP000245341 UP000248481 UP000265300 UP000286641
UP000095300 UP000008820 UP000037069 UP000069940 UP000249989 UP000000304 UP000000803 UP000091820 UP000192221 UP000092445 UP000078200 UP000092444 UP000008711 UP000095301 UP000002282 UP000007798 UP000001292 UP000008792 UP000268350 UP000008744 UP000001819 UP000092461 UP000007801 UP000092553 UP000002358 UP000001070 UP000009192 UP000215335 UP000279307 UP000235965 UP000008237 UP000092462 UP000005205 UP000005203 UP000007755 UP000053097 UP000183832 UP000007266 UP000000311 UP000027135 UP000245037 UP000242457 UP000192223 UP000076858 UP000078541 UP000078540 UP000075809 UP000078492 UP000000305 UP000230750 UP000078542 UP000076502 UP000001646 UP000242188 UP000286640 UP000002254 UP000245341 UP000248481 UP000265300 UP000286641
Pfam
Interpro
IPR007724
Poly_GlycHdrlase
+ More
IPR018862 eIF4E-T
IPR036322 WD40_repeat_dom_sf
IPR013320 ConA-like_dom_sf
IPR000306 Znf_FYVE
IPR019775 WD40_repeat_CS
IPR000409 BEACH_dom
IPR031570 DUF4704
IPR036372 BEACH_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR011011 Znf_FYVE_PHD
IPR001680 WD40_repeat
IPR017455 Znf_FYVE-rel
IPR016024 ARM-type_fold
IPR017986 WD40_repeat_dom
IPR023362 PH-BEACH_dom
IPR013083 Znf_RING/FYVE/PHD
IPR018862 eIF4E-T
IPR036322 WD40_repeat_dom_sf
IPR013320 ConA-like_dom_sf
IPR000306 Znf_FYVE
IPR019775 WD40_repeat_CS
IPR000409 BEACH_dom
IPR031570 DUF4704
IPR036372 BEACH_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR011011 Znf_FYVE_PHD
IPR001680 WD40_repeat
IPR017455 Znf_FYVE-rel
IPR016024 ARM-type_fold
IPR017986 WD40_repeat_dom
IPR023362 PH-BEACH_dom
IPR013083 Znf_RING/FYVE/PHD
SUPFAM
Gene 3D
ProteinModelPortal
H9J8P3
A0A3S2NV82
A0A2A4J6Y6
A0A194PU76
A0A212ER22
B0WST7
+ More
A0A1Q3F4V3 A0A1I8P7Y8 Q17L69 A0A0L0C4D8 A0A0A1WZR8 A0A182GTV7 B4R467 A0A212FBJ9 A0A034W105 O46043 A0A1A9W8L6 A0A1W4VSH3 A0A0K8UU75 A0A1B0ADR4 A0A1A9VY97 A0A0K8UFZ1 W8AQL3 A0A1B0FJ80 B3NSQ9 A0A1I8NDE2 B4Q0I7 A0A182GBX4 A0A1I8NDE8 U5EQC3 B4NPN3 B4I158 A0A1L8DL32 B4M1L2 A0A3B0JWQ9 B4H0U9 A0A3B0JRR4 Q29GN0 A0A1B0GHX2 B3MW84 A0A0M4F7R8 K7J0P1 B4JN37 B4L4V6 A0A0C9QWZ8 A0A232F6A4 A0A3L8DI25 A0A2J7PYB3 A0A2J7PY66 E2BI70 A0A1B0DM96 A0A2J7PY95 A0A158NCD2 A0A088AIJ7 F4WWX1 A0A1B6KY88 A0A2J7PY76 A0A336MPT7 A0A1B6HT42 A0A026VX02 A0A1B6HAF6 A0A1J1HID5 D6W990 E2B1V8 A0A067RFC3 A0A2P8Z7V8 A0A2A3EK29 A0A1W4WD06 A0A0P5Y1S9 A0A0P5Q659 A0A195FAJ8 A0A0P5NMJ0 A0A195BY71 A0A151WS21 A0A151ITL5 A0A0N8BQQ5 A0A0P5QQE3 A0A069DVA2 A0A0P6K040 A0A2S2QZC1 E9GAG4 A0A2G8K0Z3 A0A151IG65 A0A1Y1KZH8 A0A154PFY3 A0A2D4MHK1 H9GUD9 A0A210PSB1 A0A3Q7RU58 J9NTQ1 A0A2U3X9T8 A0A2U3X9S9 A0A2Y9HZM2 A0A340WI96 A0A3Q7PSD1 A0A340WBF8 A0A0F7ZDZ9 K9IUD8
A0A1Q3F4V3 A0A1I8P7Y8 Q17L69 A0A0L0C4D8 A0A0A1WZR8 A0A182GTV7 B4R467 A0A212FBJ9 A0A034W105 O46043 A0A1A9W8L6 A0A1W4VSH3 A0A0K8UU75 A0A1B0ADR4 A0A1A9VY97 A0A0K8UFZ1 W8AQL3 A0A1B0FJ80 B3NSQ9 A0A1I8NDE2 B4Q0I7 A0A182GBX4 A0A1I8NDE8 U5EQC3 B4NPN3 B4I158 A0A1L8DL32 B4M1L2 A0A3B0JWQ9 B4H0U9 A0A3B0JRR4 Q29GN0 A0A1B0GHX2 B3MW84 A0A0M4F7R8 K7J0P1 B4JN37 B4L4V6 A0A0C9QWZ8 A0A232F6A4 A0A3L8DI25 A0A2J7PYB3 A0A2J7PY66 E2BI70 A0A1B0DM96 A0A2J7PY95 A0A158NCD2 A0A088AIJ7 F4WWX1 A0A1B6KY88 A0A2J7PY76 A0A336MPT7 A0A1B6HT42 A0A026VX02 A0A1B6HAF6 A0A1J1HID5 D6W990 E2B1V8 A0A067RFC3 A0A2P8Z7V8 A0A2A3EK29 A0A1W4WD06 A0A0P5Y1S9 A0A0P5Q659 A0A195FAJ8 A0A0P5NMJ0 A0A195BY71 A0A151WS21 A0A151ITL5 A0A0N8BQQ5 A0A0P5QQE3 A0A069DVA2 A0A0P6K040 A0A2S2QZC1 E9GAG4 A0A2G8K0Z3 A0A151IG65 A0A1Y1KZH8 A0A154PFY3 A0A2D4MHK1 H9GUD9 A0A210PSB1 A0A3Q7RU58 J9NTQ1 A0A2U3X9T8 A0A2U3X9S9 A0A2Y9HZM2 A0A340WI96 A0A3Q7PSD1 A0A340WBF8 A0A0F7ZDZ9 K9IUD8
PDB
6HMN
E-value=3.83495e-97,
Score=908
Ontologies
GO
PANTHER
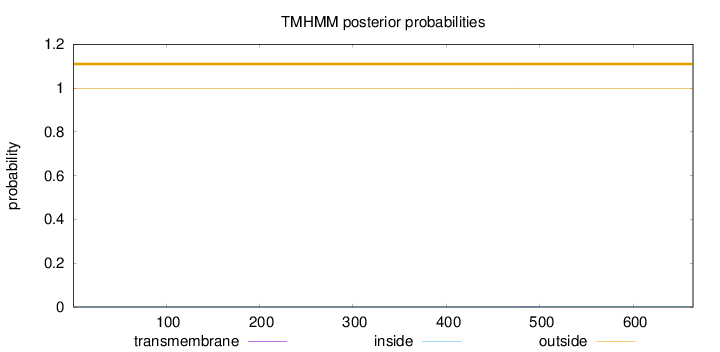
Topology
Length:
664
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02189
Exp number, first 60 AAs:
0.00061
Total prob of N-in:
0.00045
outside
1 - 664
Population Genetic Test Statistics
Pi
26.546791
Theta
24.107553
Tajima's D
0.13042
CLR
0.532711
CSRT
0.409579521023949
Interpretation
Uncertain
