Pre Gene Modal
BGIBMGA005894
Annotation
PREDICTED:_retinoblastoma-binding_protein_5_homolog_[Papilio_xuthus]
Full name
Phospholipid-transporting ATPase
+ More
Retinoblastoma-binding protein 5 homolog
Retinoblastoma-binding protein 5 homolog
Location in the cell
Cytoplasmic Reliability : 1.141 Nuclear Reliability : 1.542
Sequence
CDS
ATGAATTTAGAGTTACTGGAATCGTTTGGACAAAATTATCCTGAGGAATTCGATGGTACCCTTGACTCAATTTCACTGGCTTCAACTTGCGCATTCAACAGACGAGGTACATTGCTGGCTGTAGGCTGCAATGATGGTAGGATTTTTATCTGGGATTTCCTGACTCGGGGCATTGCAAAGTCAATATCAGCTCATGTGTACCCTGTATGCAGCTTAAGTTGGTCAAGGAATTGTAAAAAGCTATTATCAGCATCAACAGATCATAATGTATGCATATGGGATGTACTGTCTGGTGAGTGTGAACAGACGTACAGATTCCCTACTTCAATATTACGTGTGCAATTTGATCCGAGGAATGACAAGAGATTTCTCGTGTGTCCCATGAGACATGCAGCCTTACTGGTCGATACTGATGGTGATCATAAAATACTTCCTATAGACGAAGATGGAGATGTAAATGTAATGGCTTCCTTCAACAGAAGAGGAGACTATGTTTATACAGGGAATGCCAAAGGAAAAATACTGATACTTGACTCGCAGACGTTGGCCGTAAAAGCCAGTTTTAAAATAACTGTAGGCACATCCAGTACAACTAGTATCAAGAGCATCGAATTTGCTAGAAGGGGAGATTGTTTCCTAGTGAACACGTCGGACCGCGTCATCCGGGTGTACGACACCAACACGGTGCTCCAGTGCGGGGTCAACGGTGAACCCGAACCCATACAGAAACTCCAGGACCTCATCAACAAAACAACGTGGAAGAAATGTTGTTTCTCCGGCGACGGCGAGTACATCTGCGCCGGGTCCGCGCGACAGCACGCGCTCTACATCTGGGAGAAGTCGATAGGGAACCTCGTCAAGATCCTGCACGGAACCAAGGGGGAGCTCCTGCTGGACGTCGTGTGGCACCCCATCCGCCCCATCATCGCCAGCATCAGCGCCGGTGTGGTATCGATATGGGCACAGAACCAAGTCGAGAACTGGTCAGCGTTCGCTCCCGACTTCAAGGAGCTGGACGAGAACGTGGAGTACGAGGAGCGAGAGAGCGAGTTCGACGTGGAGGACGAGGACCGCTCTGTGGACCAGGCCGGGGACACCAGGGACGACGAGGAAGTCGAGGTAGACGTGACTTCGTGCGAAGCGGTGGCGGCCTTCTGCAGCTCCGACGAGGAGGGCGAGGACGAGAACGTGCTGCTGTTCCTGCCCACCGCCCCCGAGATCGAGGACCCCGAGGATGGTTGGGCGGCGACTCAGGAGACGATCACGCCAGCCGAAACTCCAGAAAAATTGGAACCTGCAGCAAAGAGGCCTAAAAGCAAAACCTACGACATATCACTGAAGATTCCCCCGCCTGAACAATCATTGTCATTCGGAGGGAAAAACAAACAAGCTGCTGGAAATAAAAAAGTAGCGGGCAGGCCGAGGAAATAA
Protein
MNLELLESFGQNYPEEFDGTLDSISLASTCAFNRRGTLLAVGCNDGRIFIWDFLTRGIAKSISAHVYPVCSLSWSRNCKKLLSASTDHNVCIWDVLSGECEQTYRFPTSILRVQFDPRNDKRFLVCPMRHAALLVDTDGDHKILPIDEDGDVNVMASFNRRGDYVYTGNAKGKILILDSQTLAVKASFKITVGTSSTTSIKSIEFARRGDCFLVNTSDRVIRVYDTNTVLQCGVNGEPEPIQKLQDLINKTTWKKCCFSGDGEYICAGSARQHALYIWEKSIGNLVKILHGTKGELLLDVVWHPIRPIIASISAGVVSIWAQNQVENWSAFAPDFKELDENVEYEERESEFDVEDEDRSVDQAGDTRDDEEVEVDVTSCEAVAAFCSSDEEGEDENVLLFLPTAPEIEDPEDGWAATQETITPAETPEKLEPAAKRPKSKTYDISLKIPPPEQSLSFGGKNKQAAGNKKVAGRPRK
Summary
Description
Component of the SET1 complex that specifically di- and trimethylates 'Lys-4' of histone H3 and of the MLL3/4 complex which also methylates histone H3 'Lys-4'.
Catalytic Activity
ATP + H(2)O + phospholipid(Side 1) = ADP + phosphate + phospholipid(Side 2).
Subunit
Core component of several methyltransferase-containing complexes. Component of the SET1 complex, composed at least of the catalytic subunit Set1, wds/WDR5, Wdr82, Rbbp5, ash2, Cfp1/CXXC1, hcf and Dpy-30L1. Component of the MLL3/4 complex composed at least of the catalytic subunit trr, ash2, Rbbp5, Dpy-30L1, wds, hcf, ptip, Pa1, Utx, Lpt and Ncoa6.
Similarity
Belongs to the cation transport ATPase (P-type) (TC 3.A.3) family. Type IV subfamily.
Keywords
Chromatin regulator
Complete proteome
Nucleus
Reference proteome
Repeat
Transcription
Transcription regulation
WD repeat
Feature
chain Retinoblastoma-binding protein 5 homolog
Uniprot
H9J8Q2
A0A194PV58
A0A2A4J7P1
A0A0N0PBM4
A0A212EYA4
A0A2H1VSE5
+ More
T1PK06 B4PFA7 A0A2J7R287 B3NIJ6 A0A0L0C3X6 A0A347ZJ83 B4QRX1 A0A1A9ZFY3 Q2M0V3 B4GR18 A0A182GZA4 W8C4P1 A0A1A9WPV6 A0A1I8PAV3 B4MLK6 Q9VPH8 Q17GA8 Q7Q8T2 B4LGF3 A0A0A1XID5 A0A3B0JRH8 A0A034VKM4 B3M974 A0A1A9V2Y3 A0A1B0ARA7 A0A1Q3F453 A0A1W4W5K5 A0A182S4X8 B0WTZ2 A0A026VXJ9 B4IWX6 A0A182JA02 A0A0M4EZK9 B4KY19 A0A182VKQ3 D6WGH2 A0A182LQ18 A0A1Y9GK61 A0A182TRM4 A0A182MYD8 A0A2M4AJL7 W5JDI7 A0A182MC05 A0A0K8WLX1 E2BCB0 A0A1Y1LIR5 F4X2E6 A0A182R137 A0A1B6G611 A0A182VTC1 N6THE0 A0A088AE91 N6UDR3 A0A1A9YL35 A0A182YA36 A0A2A3ERQ1 A0A084VKW5 A0A182X1U7 A0A0J7L8J5 A0A0C9RK96 A0A0L7R254 A0A182P8H0 A0A195CSA8 A0A151JYI6 E0VFH9 A0A1L8DNF0 A0A2R5LL22 A0A154P6C9 A0A224YQH2 A0A131YN32 A0A1S3CX87 A0A131XNU8 A0A131XZV1 L7MAM2 L7MAI9 A0A224YK23 A0A131YEG0 A0A067QXV9 A0A1E1XRD4 A0A182FUJ7 A0A1E1XF60 T1GDK4 A0A023GBU5 A0A0K8RHX0 A0A336MF23 A0A1B6MA67 K7INB4 A0A293MTH2 A0A1J1II07 A0A1B6E8W2 A0A232EYF4 A0A151WV00 A0A0P6IBQ3 A0A0P4ZRD7
T1PK06 B4PFA7 A0A2J7R287 B3NIJ6 A0A0L0C3X6 A0A347ZJ83 B4QRX1 A0A1A9ZFY3 Q2M0V3 B4GR18 A0A182GZA4 W8C4P1 A0A1A9WPV6 A0A1I8PAV3 B4MLK6 Q9VPH8 Q17GA8 Q7Q8T2 B4LGF3 A0A0A1XID5 A0A3B0JRH8 A0A034VKM4 B3M974 A0A1A9V2Y3 A0A1B0ARA7 A0A1Q3F453 A0A1W4W5K5 A0A182S4X8 B0WTZ2 A0A026VXJ9 B4IWX6 A0A182JA02 A0A0M4EZK9 B4KY19 A0A182VKQ3 D6WGH2 A0A182LQ18 A0A1Y9GK61 A0A182TRM4 A0A182MYD8 A0A2M4AJL7 W5JDI7 A0A182MC05 A0A0K8WLX1 E2BCB0 A0A1Y1LIR5 F4X2E6 A0A182R137 A0A1B6G611 A0A182VTC1 N6THE0 A0A088AE91 N6UDR3 A0A1A9YL35 A0A182YA36 A0A2A3ERQ1 A0A084VKW5 A0A182X1U7 A0A0J7L8J5 A0A0C9RK96 A0A0L7R254 A0A182P8H0 A0A195CSA8 A0A151JYI6 E0VFH9 A0A1L8DNF0 A0A2R5LL22 A0A154P6C9 A0A224YQH2 A0A131YN32 A0A1S3CX87 A0A131XNU8 A0A131XZV1 L7MAM2 L7MAI9 A0A224YK23 A0A131YEG0 A0A067QXV9 A0A1E1XRD4 A0A182FUJ7 A0A1E1XF60 T1GDK4 A0A023GBU5 A0A0K8RHX0 A0A336MF23 A0A1B6MA67 K7INB4 A0A293MTH2 A0A1J1II07 A0A1B6E8W2 A0A232EYF4 A0A151WV00 A0A0P6IBQ3 A0A0P4ZRD7
EC Number
7.6.2.1
Pubmed
19121390
26354079
22118469
25315136
17994087
17550304
+ More
26108605 22936249 15632085 26483478 24495485 10731132 12537572 12537569 21694722 21875999 17510324 12364791 25830018 25348373 24508170 18362917 19820115 20966253 20920257 23761445 20798317 28004739 21719571 23537049 25244985 24438588 20566863 28797301 26830274 28049606 25576852 24845553 29209593 28503490 20075255 28648823
26108605 22936249 15632085 26483478 24495485 10731132 12537572 12537569 21694722 21875999 17510324 12364791 25830018 25348373 24508170 18362917 19820115 20966253 20920257 23761445 20798317 28004739 21719571 23537049 25244985 24438588 20566863 28797301 26830274 28049606 25576852 24845553 29209593 28503490 20075255 28648823
EMBL
BABH01030362
BABH01030363
KQ459592
KPI96873.1
NWSH01002597
PCG67889.1
+ More
KQ460954 KPJ10256.1 AGBW02011576 OWR46472.1 ODYU01004162 SOQ43739.1 KA648999 AFP63628.1 CM000159 EDW95193.1 NEVH01008200 PNF34932.1 CH954178 EDV52492.1 JRES01000936 KNC27053.1 FX985651 BBA84389.1 CM000363 CM002912 EDX11217.1 KMZ00744.1 CH379069 EAL30823.2 CH479188 EDW40203.1 JXUM01099203 KQ564476 KXJ72222.1 GAMC01005379 JAC01177.1 CH963847 EDW72862.1 AE014296 AY071120 BT032759 BT032765 AAF51573.2 AAL48742.1 ACD81773.1 CH477264 EAT45597.1 AAAB01008933 EAA09974.3 CH940647 EDW70482.1 GBXI01004044 JAD10248.1 OUUW01000009 SPP84735.1 GAKP01015918 JAC43034.1 CH902618 EDV40058.2 JXJN01002301 GFDL01012711 JAV22334.1 DS232096 EDS34699.1 KK107801 EZA47584.1 CH916366 EDV97377.1 CP012525 ALC44162.1 CH933809 EDW18721.1 KQ971321 EFA00179.1 APCN01005566 GGFK01007640 MBW40961.1 ADMH02001479 ETN62412.1 AXCM01002293 GDHF01000173 JAI52141.1 GL447257 EFN86646.1 GEZM01060786 JAV70917.1 GL888578 EGI59371.1 AXCN02001048 GECZ01011893 JAS57876.1 APGK01026531 KB740635 ENN79844.1 KB631992 ENN79845.1 ERL87749.1 KZ288192 PBC34370.1 ATLV01014310 KE524956 KFB38609.1 LBMM01000267 KMR02294.1 GBYB01007256 GBYB01007259 JAG77023.1 JAG77026.1 KQ414666 KOC64908.1 KQ977329 KYN03530.1 KQ981477 KYN41447.1 DS235112 EEB12135.1 GFDF01006143 JAV07941.1 GGLE01006100 MBY10226.1 KQ434827 KZC07496.1 GFPF01006863 MAA18009.1 GEDV01008579 JAP79978.1 GEFH01000686 JAP67895.1 GEFM01003033 JAP72763.1 GACK01004865 JAA60169.1 GACK01004866 JAA60168.1 GFPF01006862 MAA18008.1 GEDV01011707 JAP76850.1 KK852832 KDR15310.1 GFAA01001566 JAU01869.1 GFAC01001308 JAT97880.1 CAQQ02074621 GBBM01003727 JAC31691.1 GADI01003088 JAA70720.1 UFQT01001098 SSX28905.1 GEBQ01025684 GEBQ01007170 JAT14293.1 JAT32807.1 GFWV01019406 MAA44134.1 CVRI01000048 CRK98686.1 GEDC01002926 JAS34372.1 NNAY01001642 OXU23339.1 KQ982721 KYQ51666.1 GDIQ01023258 JAN71479.1 GDIP01213398 JAJ10004.1
KQ460954 KPJ10256.1 AGBW02011576 OWR46472.1 ODYU01004162 SOQ43739.1 KA648999 AFP63628.1 CM000159 EDW95193.1 NEVH01008200 PNF34932.1 CH954178 EDV52492.1 JRES01000936 KNC27053.1 FX985651 BBA84389.1 CM000363 CM002912 EDX11217.1 KMZ00744.1 CH379069 EAL30823.2 CH479188 EDW40203.1 JXUM01099203 KQ564476 KXJ72222.1 GAMC01005379 JAC01177.1 CH963847 EDW72862.1 AE014296 AY071120 BT032759 BT032765 AAF51573.2 AAL48742.1 ACD81773.1 CH477264 EAT45597.1 AAAB01008933 EAA09974.3 CH940647 EDW70482.1 GBXI01004044 JAD10248.1 OUUW01000009 SPP84735.1 GAKP01015918 JAC43034.1 CH902618 EDV40058.2 JXJN01002301 GFDL01012711 JAV22334.1 DS232096 EDS34699.1 KK107801 EZA47584.1 CH916366 EDV97377.1 CP012525 ALC44162.1 CH933809 EDW18721.1 KQ971321 EFA00179.1 APCN01005566 GGFK01007640 MBW40961.1 ADMH02001479 ETN62412.1 AXCM01002293 GDHF01000173 JAI52141.1 GL447257 EFN86646.1 GEZM01060786 JAV70917.1 GL888578 EGI59371.1 AXCN02001048 GECZ01011893 JAS57876.1 APGK01026531 KB740635 ENN79844.1 KB631992 ENN79845.1 ERL87749.1 KZ288192 PBC34370.1 ATLV01014310 KE524956 KFB38609.1 LBMM01000267 KMR02294.1 GBYB01007256 GBYB01007259 JAG77023.1 JAG77026.1 KQ414666 KOC64908.1 KQ977329 KYN03530.1 KQ981477 KYN41447.1 DS235112 EEB12135.1 GFDF01006143 JAV07941.1 GGLE01006100 MBY10226.1 KQ434827 KZC07496.1 GFPF01006863 MAA18009.1 GEDV01008579 JAP79978.1 GEFH01000686 JAP67895.1 GEFM01003033 JAP72763.1 GACK01004865 JAA60169.1 GACK01004866 JAA60168.1 GFPF01006862 MAA18008.1 GEDV01011707 JAP76850.1 KK852832 KDR15310.1 GFAA01001566 JAU01869.1 GFAC01001308 JAT97880.1 CAQQ02074621 GBBM01003727 JAC31691.1 GADI01003088 JAA70720.1 UFQT01001098 SSX28905.1 GEBQ01025684 GEBQ01007170 JAT14293.1 JAT32807.1 GFWV01019406 MAA44134.1 CVRI01000048 CRK98686.1 GEDC01002926 JAS34372.1 NNAY01001642 OXU23339.1 KQ982721 KYQ51666.1 GDIQ01023258 JAN71479.1 GDIP01213398 JAJ10004.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000007151
UP000095301
+ More
UP000002282 UP000235965 UP000008711 UP000037069 UP000000304 UP000092445 UP000001819 UP000008744 UP000069940 UP000249989 UP000091820 UP000095300 UP000007798 UP000000803 UP000008820 UP000007062 UP000008792 UP000268350 UP000007801 UP000078200 UP000092460 UP000192221 UP000075900 UP000002320 UP000053097 UP000001070 UP000075880 UP000092553 UP000009192 UP000075903 UP000007266 UP000075882 UP000075840 UP000075902 UP000075884 UP000000673 UP000075883 UP000008237 UP000007755 UP000075886 UP000075920 UP000019118 UP000005203 UP000030742 UP000092443 UP000076408 UP000242457 UP000030765 UP000076407 UP000036403 UP000053825 UP000075885 UP000078542 UP000078541 UP000009046 UP000076502 UP000079169 UP000027135 UP000069272 UP000015102 UP000002358 UP000183832 UP000215335 UP000075809
UP000002282 UP000235965 UP000008711 UP000037069 UP000000304 UP000092445 UP000001819 UP000008744 UP000069940 UP000249989 UP000091820 UP000095300 UP000007798 UP000000803 UP000008820 UP000007062 UP000008792 UP000268350 UP000007801 UP000078200 UP000092460 UP000192221 UP000075900 UP000002320 UP000053097 UP000001070 UP000075880 UP000092553 UP000009192 UP000075903 UP000007266 UP000075882 UP000075840 UP000075902 UP000075884 UP000000673 UP000075883 UP000008237 UP000007755 UP000075886 UP000075920 UP000019118 UP000005203 UP000030742 UP000092443 UP000076408 UP000242457 UP000030765 UP000076407 UP000036403 UP000053825 UP000075885 UP000078542 UP000078541 UP000009046 UP000076502 UP000079169 UP000027135 UP000069272 UP000015102 UP000002358 UP000183832 UP000215335 UP000075809
Pfam
Interpro
IPR015943
WD40/YVTN_repeat-like_dom_sf
+ More
IPR001680 WD40_repeat
IPR036361 SAP_dom_sf
IPR017986 WD40_repeat_dom
IPR003034 SAP_dom
IPR036322 WD40_repeat_dom_sf
IPR037850 RBBP5/Swd1
IPR019775 WD40_repeat_CS
IPR032630 P_typ_ATPase_c
IPR018303 ATPase_P-typ_P_site
IPR008250 ATPase_P-typ_transduc_dom_A_sf
IPR023298 ATPase_P-typ_TM_dom_sf
IPR001757 P_typ_ATPase
IPR023299 ATPase_P-typ_cyto_dom_N
IPR006539 P-type_ATPase_IV
IPR032631 P-type_ATPase_N
IPR036412 HAD-like_sf
IPR023214 HAD_sf
IPR011990 TPR-like_helical_dom_sf
IPR016024 ARM-type_fold
IPR024679 Ipi1_N
IPR019410 Methyltransf_16
IPR029063 SAM-dependent_MTases
IPR029426 FAM86
IPR013594 Dynein_heavy_dom-1
IPR001007 VWF_dom
IPR003961 FN3_dom
IPR013783 Ig-like_fold
IPR036116 FN3_sf
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR001680 WD40_repeat
IPR036361 SAP_dom_sf
IPR017986 WD40_repeat_dom
IPR003034 SAP_dom
IPR036322 WD40_repeat_dom_sf
IPR037850 RBBP5/Swd1
IPR019775 WD40_repeat_CS
IPR032630 P_typ_ATPase_c
IPR018303 ATPase_P-typ_P_site
IPR008250 ATPase_P-typ_transduc_dom_A_sf
IPR023298 ATPase_P-typ_TM_dom_sf
IPR001757 P_typ_ATPase
IPR023299 ATPase_P-typ_cyto_dom_N
IPR006539 P-type_ATPase_IV
IPR032631 P-type_ATPase_N
IPR036412 HAD-like_sf
IPR023214 HAD_sf
IPR011990 TPR-like_helical_dom_sf
IPR016024 ARM-type_fold
IPR024679 Ipi1_N
IPR019410 Methyltransf_16
IPR029063 SAM-dependent_MTases
IPR029426 FAM86
IPR013594 Dynein_heavy_dom-1
IPR001007 VWF_dom
IPR003961 FN3_dom
IPR013783 Ig-like_fold
IPR036116 FN3_sf
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
SUPFAM
CDD
ProteinModelPortal
H9J8Q2
A0A194PV58
A0A2A4J7P1
A0A0N0PBM4
A0A212EYA4
A0A2H1VSE5
+ More
T1PK06 B4PFA7 A0A2J7R287 B3NIJ6 A0A0L0C3X6 A0A347ZJ83 B4QRX1 A0A1A9ZFY3 Q2M0V3 B4GR18 A0A182GZA4 W8C4P1 A0A1A9WPV6 A0A1I8PAV3 B4MLK6 Q9VPH8 Q17GA8 Q7Q8T2 B4LGF3 A0A0A1XID5 A0A3B0JRH8 A0A034VKM4 B3M974 A0A1A9V2Y3 A0A1B0ARA7 A0A1Q3F453 A0A1W4W5K5 A0A182S4X8 B0WTZ2 A0A026VXJ9 B4IWX6 A0A182JA02 A0A0M4EZK9 B4KY19 A0A182VKQ3 D6WGH2 A0A182LQ18 A0A1Y9GK61 A0A182TRM4 A0A182MYD8 A0A2M4AJL7 W5JDI7 A0A182MC05 A0A0K8WLX1 E2BCB0 A0A1Y1LIR5 F4X2E6 A0A182R137 A0A1B6G611 A0A182VTC1 N6THE0 A0A088AE91 N6UDR3 A0A1A9YL35 A0A182YA36 A0A2A3ERQ1 A0A084VKW5 A0A182X1U7 A0A0J7L8J5 A0A0C9RK96 A0A0L7R254 A0A182P8H0 A0A195CSA8 A0A151JYI6 E0VFH9 A0A1L8DNF0 A0A2R5LL22 A0A154P6C9 A0A224YQH2 A0A131YN32 A0A1S3CX87 A0A131XNU8 A0A131XZV1 L7MAM2 L7MAI9 A0A224YK23 A0A131YEG0 A0A067QXV9 A0A1E1XRD4 A0A182FUJ7 A0A1E1XF60 T1GDK4 A0A023GBU5 A0A0K8RHX0 A0A336MF23 A0A1B6MA67 K7INB4 A0A293MTH2 A0A1J1II07 A0A1B6E8W2 A0A232EYF4 A0A151WV00 A0A0P6IBQ3 A0A0P4ZRD7
T1PK06 B4PFA7 A0A2J7R287 B3NIJ6 A0A0L0C3X6 A0A347ZJ83 B4QRX1 A0A1A9ZFY3 Q2M0V3 B4GR18 A0A182GZA4 W8C4P1 A0A1A9WPV6 A0A1I8PAV3 B4MLK6 Q9VPH8 Q17GA8 Q7Q8T2 B4LGF3 A0A0A1XID5 A0A3B0JRH8 A0A034VKM4 B3M974 A0A1A9V2Y3 A0A1B0ARA7 A0A1Q3F453 A0A1W4W5K5 A0A182S4X8 B0WTZ2 A0A026VXJ9 B4IWX6 A0A182JA02 A0A0M4EZK9 B4KY19 A0A182VKQ3 D6WGH2 A0A182LQ18 A0A1Y9GK61 A0A182TRM4 A0A182MYD8 A0A2M4AJL7 W5JDI7 A0A182MC05 A0A0K8WLX1 E2BCB0 A0A1Y1LIR5 F4X2E6 A0A182R137 A0A1B6G611 A0A182VTC1 N6THE0 A0A088AE91 N6UDR3 A0A1A9YL35 A0A182YA36 A0A2A3ERQ1 A0A084VKW5 A0A182X1U7 A0A0J7L8J5 A0A0C9RK96 A0A0L7R254 A0A182P8H0 A0A195CSA8 A0A151JYI6 E0VFH9 A0A1L8DNF0 A0A2R5LL22 A0A154P6C9 A0A224YQH2 A0A131YN32 A0A1S3CX87 A0A131XNU8 A0A131XZV1 L7MAM2 L7MAI9 A0A224YK23 A0A131YEG0 A0A067QXV9 A0A1E1XRD4 A0A182FUJ7 A0A1E1XF60 T1GDK4 A0A023GBU5 A0A0K8RHX0 A0A336MF23 A0A1B6MA67 K7INB4 A0A293MTH2 A0A1J1II07 A0A1B6E8W2 A0A232EYF4 A0A151WV00 A0A0P6IBQ3 A0A0P4ZRD7
PDB
5OV3
E-value=6.14732e-149,
Score=1353
Ontologies
GO
Topology
Subcellular location
Membrane
Nucleus
Nucleus
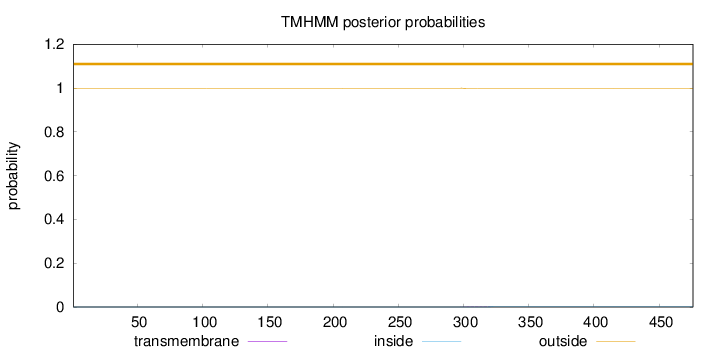
Length:
476
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0355
Exp number, first 60 AAs:
0.00068
Total prob of N-in:
0.00042
outside
1 - 476
Population Genetic Test Statistics
Pi
19.744297
Theta
18.39739
Tajima's D
-0.631767
CLR
0.587368
CSRT
0.215839208039598
Interpretation
Uncertain
