Pre Gene Modal
BGIBMGA005895
Annotation
PREDICTED:_uncharacterized_protein_CG5902_[Papilio_machaon]
Location in the cell
Mitochondrial Reliability : 3.022
Sequence
CDS
ATGGAACCACCGCAGACGCCCAAATTCACAAACGAGGCCTATCCTTTATTTGTGACTTGGAAGATCGGGAAAGAGCACCGTCTAAGGGGATGCATTGGGACCTTCAACGCTATGCACTTACATTCAGGCCTGCGTGAATATGCTATTACAAGTGCTCTGAGAGACTCTCGATTTTCTCCTATAACAAGAGAGGAAGTTCCGCGGCTCTCTGTCTCGGTGTCAATCCTGCAGCATTTTGAAGAAGCTGAACATTACCTTGATTGGAAACTGGGCAAGCATGGCATCCGCATAGAATTTGTCAGTGAACGTGGAAGCAAACGTACGGCTACATACTTACCTCAAGTTGCTACTGAGCAAGGTTGGGACCAAATACAAACAATAGATTCTTTACTAAGGAAGGGAGGTTACAAAGCTGCAATAACAGCAGATTTGCGACGCAGCATCAAACTAACTCGATATCAGTCTGAGGAGGTTTCCGCCTCATACACTGATTATGTGAACCGGCGTTGCTAA
Protein
MEPPQTPKFTNEAYPLFVTWKIGKEHRLRGCIGTFNAMHLHSGLREYAITSALRDSRFSPITREEVPRLSVSVSILQHFEEAEHYLDWKLGKHGIRIEFVSERGSKRTATYLPQVATEQGWDQIQTIDSLLRKGGYKAAITADLRRSIKLTRYQSEEVSASYTDYVNRRC
Summary
Uniprot
H9J8Q3
A0A0N1I5V8
A0A194PTY6
A0A1E1WQX1
S4PKE4
A0A2A4JHL0
+ More
A0A212EYB3 A0A2H1VSE2 A0A0L7LJJ9 A0A3S2P116 A0A1Y1MQW1 A0A0T6BG66 N6SWH6 D7EIE8 A0A1W4W4X5 A0A0J7L8Z5 A0A151J5L8 F4X203 A0A195FJN2 A0A026W6S9 E2AQC4 A0A2J7PHD2 A0A158P3R7 A0A2P8Z259 A0A158P3R6 A0A1I8M7T2 A0A0K8TPT0 A0A195FJC2 Q16X97 A0A182MCV8 A0A182GV63 A0A1I8Q2M7 A0A182W566 A0A034VVK2 A0A0K8WKB3 E0W1J5 D3TNR7 A0A1B0BX11 A0A0A1WLC9 A0A1L8DB25 A0A182P8J2 A0A034VS41 A0A1A9UM65 A0A0J7L9S5 A0A154P073 A0A1Q3EZY0 A0A1Q3EZY6 B0W9F9 A0A182VGW5 A0A1B6E9M0 A0A2L2YI97 W8BG67 A0A182RM43 W5JJG3 A0A3L8DW39 A0A182FK83 A0A226E399 A0A2M4AN00 A0A2M4AN46 A0A1B6F9P2 A0A1B6IGS7 A0A2M4AN41 A0A2M4AN50 A0A2M4BVI0 A0A2M4BV90 A0A2A3ELH0 A0A087ZPG5 A0A182LM50 Q7QF70 A0A3F2Z202 A0A182HMX1 A0A2M3Z7A1 A0A2M4BWX0 A0A023GAH0 G3MMI0 A0A1E1XS80 A0A023FZE0 A0A182JDG3 A0A1W4UL62 A0A1D2NJG6 A0A087SZ47 A0A1E1X4J4 A0A182MZJ7 A0A084VQ69 A0A023FQ23 A0A1Z5LGE2 B4N9E1 A0A182KE40 A0A131XBC2 A0A0L7QZQ8 A0A224YNI8 A0A131YPL4 L7M814 E2C7K5 B3M309 A0A1B0G5S1 B4K6V0 A0A182QWH8 B4M0H0 R7U7Z3
A0A212EYB3 A0A2H1VSE2 A0A0L7LJJ9 A0A3S2P116 A0A1Y1MQW1 A0A0T6BG66 N6SWH6 D7EIE8 A0A1W4W4X5 A0A0J7L8Z5 A0A151J5L8 F4X203 A0A195FJN2 A0A026W6S9 E2AQC4 A0A2J7PHD2 A0A158P3R7 A0A2P8Z259 A0A158P3R6 A0A1I8M7T2 A0A0K8TPT0 A0A195FJC2 Q16X97 A0A182MCV8 A0A182GV63 A0A1I8Q2M7 A0A182W566 A0A034VVK2 A0A0K8WKB3 E0W1J5 D3TNR7 A0A1B0BX11 A0A0A1WLC9 A0A1L8DB25 A0A182P8J2 A0A034VS41 A0A1A9UM65 A0A0J7L9S5 A0A154P073 A0A1Q3EZY0 A0A1Q3EZY6 B0W9F9 A0A182VGW5 A0A1B6E9M0 A0A2L2YI97 W8BG67 A0A182RM43 W5JJG3 A0A3L8DW39 A0A182FK83 A0A226E399 A0A2M4AN00 A0A2M4AN46 A0A1B6F9P2 A0A1B6IGS7 A0A2M4AN41 A0A2M4AN50 A0A2M4BVI0 A0A2M4BV90 A0A2A3ELH0 A0A087ZPG5 A0A182LM50 Q7QF70 A0A3F2Z202 A0A182HMX1 A0A2M3Z7A1 A0A2M4BWX0 A0A023GAH0 G3MMI0 A0A1E1XS80 A0A023FZE0 A0A182JDG3 A0A1W4UL62 A0A1D2NJG6 A0A087SZ47 A0A1E1X4J4 A0A182MZJ7 A0A084VQ69 A0A023FQ23 A0A1Z5LGE2 B4N9E1 A0A182KE40 A0A131XBC2 A0A0L7QZQ8 A0A224YNI8 A0A131YPL4 L7M814 E2C7K5 B3M309 A0A1B0G5S1 B4K6V0 A0A182QWH8 B4M0H0 R7U7Z3
Pubmed
19121390
26354079
23622113
22118469
26227816
28004739
+ More
23537049 18362917 19820115 21719571 24508170 30249741 20798317 21347285 29403074 25315136 26369729 17510324 26483478 25348373 20566863 20353571 25830018 26561354 24495485 20920257 23761445 20966253 12364791 14747013 17210077 22216098 29209593 27289101 28503490 24438588 28528879 17994087 28049606 28797301 26830274 25576852 18057021 23254933
23537049 18362917 19820115 21719571 24508170 30249741 20798317 21347285 29403074 25315136 26369729 17510324 26483478 25348373 20566863 20353571 25830018 26561354 24495485 20920257 23761445 20966253 12364791 14747013 17210077 22216098 29209593 27289101 28503490 24438588 28528879 17994087 28049606 28797301 26830274 25576852 18057021 23254933
EMBL
BABH01030369
KQ460954
KPJ10259.1
KQ459592
KPI96876.1
GDQN01001621
+ More
JAT89433.1 GAIX01004545 JAA88015.1 NWSH01001496 PCG71074.1 AGBW02011576 OWR46470.1 ODYU01004162 SOQ43737.1 JTDY01000838 KOB75718.1 RSAL01000060 RVE49639.1 GEZM01026918 JAV86895.1 LJIG01000617 KRT86323.1 APGK01019841 APGK01053910 KB741239 KB740134 KB632275 ENN72119.1 ENN81273.1 ERL91093.1 DS497678 EFA11903.1 LBMM01000168 KMR04651.1 KQ979970 KYN18411.1 GL888558 EGI59539.1 KQ981523 KYN40467.1 KK107373 QOIP01000004 EZA51678.1 RLU23995.1 GL441722 EFN64367.1 NEVH01025139 PNF15746.1 ADTU01008383 PYGN01000233 PSN50577.1 ADTU01008381 ADTU01008382 GDAI01001472 JAI16131.1 KYN40466.1 CH477545 EAT39221.1 AXCM01000050 JXUM01059616 JXUM01090413 KQ563838 KQ562062 KXJ73192.1 KXJ76770.1 GAKP01012830 GAKP01012829 GAKP01012828 JAC46122.1 GDHF01001029 JAI51285.1 DS235871 EEB19501.1 EZ423069 ADD19345.1 JXJN01022027 GBXI01014620 GBXI01007790 JAC99671.1 JAD06502.1 GFDF01010425 JAV03659.1 GAKP01012831 JAC46121.1 KMR04652.1 KQ434787 KZC05267.1 GFDL01014185 JAV20860.1 GFDL01014184 JAV20861.1 DS231864 EDS40184.1 GEDC01002672 JAS34626.1 IAAA01025616 LAA07853.1 GAMC01017936 GAMC01017935 GAMC01017934 GAMC01017933 JAB88619.1 ADMH02000984 ETN64517.1 RLU23998.1 LNIX01000007 OXA52215.1 GGFK01008842 MBW42163.1 GGFK01008879 MBW42200.1 GECZ01022842 JAS46927.1 GECU01021591 JAS86115.1 GGFK01008878 MBW42199.1 GGFK01008880 MBW42201.1 GGFJ01007861 MBW57002.1 GGFJ01007846 MBW56987.1 KZ288221 PBC32122.1 AAAB01008846 EAA06185.4 APCN01004923 GGFM01003633 MBW24384.1 GGFJ01008418 MBW57559.1 GBBM01005525 JAC29893.1 JO843081 AEO34698.1 GFAA01001300 JAU02135.1 GBBL01000466 JAC26854.1 LJIJ01000023 ODN05391.1 KK112629 KFM58136.1 GFAC01004999 JAT94189.1 ATLV01015142 KE525003 KFB40113.1 GBBK01001078 JAC23404.1 GFJQ02000504 JAW06466.1 CH964232 EDW80574.1 GEFH01004362 JAP64219.1 KQ414679 KOC64042.1 GFPF01004657 MAA15803.1 GEDV01008177 JAP80380.1 GACK01005731 JAA59303.1 GL453383 EFN76081.1 CH902617 EDV42409.1 KPU79709.1 CCAG010015751 CH933806 EDW14216.1 KRG00922.1 AXCN02001284 CH940650 EDW68349.1 KRF83790.1 AMQN01009006 KB304376 ELU02099.1
JAT89433.1 GAIX01004545 JAA88015.1 NWSH01001496 PCG71074.1 AGBW02011576 OWR46470.1 ODYU01004162 SOQ43737.1 JTDY01000838 KOB75718.1 RSAL01000060 RVE49639.1 GEZM01026918 JAV86895.1 LJIG01000617 KRT86323.1 APGK01019841 APGK01053910 KB741239 KB740134 KB632275 ENN72119.1 ENN81273.1 ERL91093.1 DS497678 EFA11903.1 LBMM01000168 KMR04651.1 KQ979970 KYN18411.1 GL888558 EGI59539.1 KQ981523 KYN40467.1 KK107373 QOIP01000004 EZA51678.1 RLU23995.1 GL441722 EFN64367.1 NEVH01025139 PNF15746.1 ADTU01008383 PYGN01000233 PSN50577.1 ADTU01008381 ADTU01008382 GDAI01001472 JAI16131.1 KYN40466.1 CH477545 EAT39221.1 AXCM01000050 JXUM01059616 JXUM01090413 KQ563838 KQ562062 KXJ73192.1 KXJ76770.1 GAKP01012830 GAKP01012829 GAKP01012828 JAC46122.1 GDHF01001029 JAI51285.1 DS235871 EEB19501.1 EZ423069 ADD19345.1 JXJN01022027 GBXI01014620 GBXI01007790 JAC99671.1 JAD06502.1 GFDF01010425 JAV03659.1 GAKP01012831 JAC46121.1 KMR04652.1 KQ434787 KZC05267.1 GFDL01014185 JAV20860.1 GFDL01014184 JAV20861.1 DS231864 EDS40184.1 GEDC01002672 JAS34626.1 IAAA01025616 LAA07853.1 GAMC01017936 GAMC01017935 GAMC01017934 GAMC01017933 JAB88619.1 ADMH02000984 ETN64517.1 RLU23998.1 LNIX01000007 OXA52215.1 GGFK01008842 MBW42163.1 GGFK01008879 MBW42200.1 GECZ01022842 JAS46927.1 GECU01021591 JAS86115.1 GGFK01008878 MBW42199.1 GGFK01008880 MBW42201.1 GGFJ01007861 MBW57002.1 GGFJ01007846 MBW56987.1 KZ288221 PBC32122.1 AAAB01008846 EAA06185.4 APCN01004923 GGFM01003633 MBW24384.1 GGFJ01008418 MBW57559.1 GBBM01005525 JAC29893.1 JO843081 AEO34698.1 GFAA01001300 JAU02135.1 GBBL01000466 JAC26854.1 LJIJ01000023 ODN05391.1 KK112629 KFM58136.1 GFAC01004999 JAT94189.1 ATLV01015142 KE525003 KFB40113.1 GBBK01001078 JAC23404.1 GFJQ02000504 JAW06466.1 CH964232 EDW80574.1 GEFH01004362 JAP64219.1 KQ414679 KOC64042.1 GFPF01004657 MAA15803.1 GEDV01008177 JAP80380.1 GACK01005731 JAA59303.1 GL453383 EFN76081.1 CH902617 EDV42409.1 KPU79709.1 CCAG010015751 CH933806 EDW14216.1 KRG00922.1 AXCN02001284 CH940650 EDW68349.1 KRF83790.1 AMQN01009006 KB304376 ELU02099.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007151
UP000037510
+ More
UP000283053 UP000019118 UP000030742 UP000007266 UP000192223 UP000036403 UP000078492 UP000007755 UP000078541 UP000053097 UP000279307 UP000000311 UP000235965 UP000005205 UP000245037 UP000095301 UP000008820 UP000075883 UP000069940 UP000249989 UP000095300 UP000075920 UP000009046 UP000092460 UP000075885 UP000078200 UP000076502 UP000002320 UP000075903 UP000075900 UP000000673 UP000069272 UP000198287 UP000242457 UP000005203 UP000075882 UP000007062 UP000076407 UP000075840 UP000075880 UP000192221 UP000094527 UP000054359 UP000075884 UP000030765 UP000007798 UP000075881 UP000053825 UP000008237 UP000007801 UP000092444 UP000009192 UP000075886 UP000008792 UP000014760
UP000283053 UP000019118 UP000030742 UP000007266 UP000192223 UP000036403 UP000078492 UP000007755 UP000078541 UP000053097 UP000279307 UP000000311 UP000235965 UP000005205 UP000245037 UP000095301 UP000008820 UP000075883 UP000069940 UP000249989 UP000095300 UP000075920 UP000009046 UP000092460 UP000075885 UP000078200 UP000076502 UP000002320 UP000075903 UP000075900 UP000000673 UP000069272 UP000198287 UP000242457 UP000005203 UP000075882 UP000007062 UP000076407 UP000075840 UP000075880 UP000192221 UP000094527 UP000054359 UP000075884 UP000030765 UP000007798 UP000075881 UP000053825 UP000008237 UP000007801 UP000092444 UP000009192 UP000075886 UP000008792 UP000014760
PRIDE
Interpro
SUPFAM
SSF143447
SSF143447
Gene 3D
ProteinModelPortal
H9J8Q3
A0A0N1I5V8
A0A194PTY6
A0A1E1WQX1
S4PKE4
A0A2A4JHL0
+ More
A0A212EYB3 A0A2H1VSE2 A0A0L7LJJ9 A0A3S2P116 A0A1Y1MQW1 A0A0T6BG66 N6SWH6 D7EIE8 A0A1W4W4X5 A0A0J7L8Z5 A0A151J5L8 F4X203 A0A195FJN2 A0A026W6S9 E2AQC4 A0A2J7PHD2 A0A158P3R7 A0A2P8Z259 A0A158P3R6 A0A1I8M7T2 A0A0K8TPT0 A0A195FJC2 Q16X97 A0A182MCV8 A0A182GV63 A0A1I8Q2M7 A0A182W566 A0A034VVK2 A0A0K8WKB3 E0W1J5 D3TNR7 A0A1B0BX11 A0A0A1WLC9 A0A1L8DB25 A0A182P8J2 A0A034VS41 A0A1A9UM65 A0A0J7L9S5 A0A154P073 A0A1Q3EZY0 A0A1Q3EZY6 B0W9F9 A0A182VGW5 A0A1B6E9M0 A0A2L2YI97 W8BG67 A0A182RM43 W5JJG3 A0A3L8DW39 A0A182FK83 A0A226E399 A0A2M4AN00 A0A2M4AN46 A0A1B6F9P2 A0A1B6IGS7 A0A2M4AN41 A0A2M4AN50 A0A2M4BVI0 A0A2M4BV90 A0A2A3ELH0 A0A087ZPG5 A0A182LM50 Q7QF70 A0A3F2Z202 A0A182HMX1 A0A2M3Z7A1 A0A2M4BWX0 A0A023GAH0 G3MMI0 A0A1E1XS80 A0A023FZE0 A0A182JDG3 A0A1W4UL62 A0A1D2NJG6 A0A087SZ47 A0A1E1X4J4 A0A182MZJ7 A0A084VQ69 A0A023FQ23 A0A1Z5LGE2 B4N9E1 A0A182KE40 A0A131XBC2 A0A0L7QZQ8 A0A224YNI8 A0A131YPL4 L7M814 E2C7K5 B3M309 A0A1B0G5S1 B4K6V0 A0A182QWH8 B4M0H0 R7U7Z3
A0A212EYB3 A0A2H1VSE2 A0A0L7LJJ9 A0A3S2P116 A0A1Y1MQW1 A0A0T6BG66 N6SWH6 D7EIE8 A0A1W4W4X5 A0A0J7L8Z5 A0A151J5L8 F4X203 A0A195FJN2 A0A026W6S9 E2AQC4 A0A2J7PHD2 A0A158P3R7 A0A2P8Z259 A0A158P3R6 A0A1I8M7T2 A0A0K8TPT0 A0A195FJC2 Q16X97 A0A182MCV8 A0A182GV63 A0A1I8Q2M7 A0A182W566 A0A034VVK2 A0A0K8WKB3 E0W1J5 D3TNR7 A0A1B0BX11 A0A0A1WLC9 A0A1L8DB25 A0A182P8J2 A0A034VS41 A0A1A9UM65 A0A0J7L9S5 A0A154P073 A0A1Q3EZY0 A0A1Q3EZY6 B0W9F9 A0A182VGW5 A0A1B6E9M0 A0A2L2YI97 W8BG67 A0A182RM43 W5JJG3 A0A3L8DW39 A0A182FK83 A0A226E399 A0A2M4AN00 A0A2M4AN46 A0A1B6F9P2 A0A1B6IGS7 A0A2M4AN41 A0A2M4AN50 A0A2M4BVI0 A0A2M4BV90 A0A2A3ELH0 A0A087ZPG5 A0A182LM50 Q7QF70 A0A3F2Z202 A0A182HMX1 A0A2M3Z7A1 A0A2M4BWX0 A0A023GAH0 G3MMI0 A0A1E1XS80 A0A023FZE0 A0A182JDG3 A0A1W4UL62 A0A1D2NJG6 A0A087SZ47 A0A1E1X4J4 A0A182MZJ7 A0A084VQ69 A0A023FQ23 A0A1Z5LGE2 B4N9E1 A0A182KE40 A0A131XBC2 A0A0L7QZQ8 A0A224YNI8 A0A131YPL4 L7M814 E2C7K5 B3M309 A0A1B0G5S1 B4K6V0 A0A182QWH8 B4M0H0 R7U7Z3
PDB
1WSC
E-value=2.79877e-08,
Score=134
Ontologies
GO
PANTHER
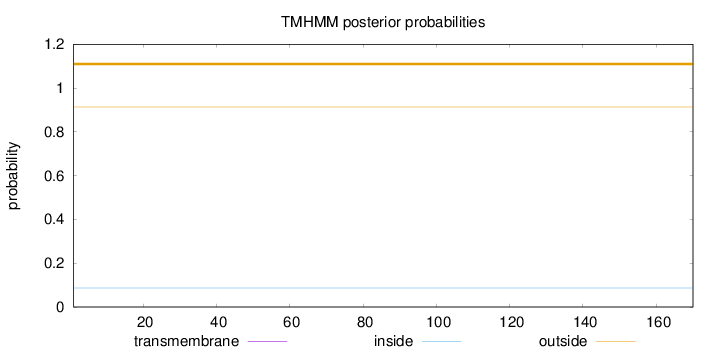
Topology
Length:
170
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00199
Exp number, first 60 AAs:
0.00199
Total prob of N-in:
0.08706
outside
1 - 170
Population Genetic Test Statistics
Pi
17.623474
Theta
13.538569
Tajima's D
0.755859
CLR
0.416465
CSRT
0.59962001899905
Interpretation
Uncertain
