Gene
KWMTBOMO07513
Pre Gene Modal
BGIBMGA005880
Annotation
PREDICTED:_probable_phospholipid-transporting_ATPase_IF_isoform_X1_[Bombyx_mori]
Full name
Phospholipid-transporting ATPase
Location in the cell
PlasmaMembrane Reliability : 4.587
Sequence
CDS
ATGGTTTCATCTGATAGTCCTGTATCGCCTTTCACAAGCATCGCTCCGTTATCATTCATGGTGTTCGTAACAGCTGTCAAGCAAGGCTACGAAGACTGGCTCAGGCACAAGACTGATAATAAAGTCAACAATCAGATTGTTGAAATAGTCTACAAAGGCGTGATCAAAGAAGTGAAAAATTCCACGATAGCCCCTGGAACCTTGGTTCGTGTAAAACGAGGAAGGGAAGTACCAGCCGACCTCGTGCTCCTCTGTTCCGCCGGAGAGAAGGGAAAATGTTATATCACCACTGCGAATCTGGACGGGGAGACCAATTTGAAAACACTAAGGGTGCCCCCACCTTTAGTCGGGTATACGCCGGACATTTTGCCACAAAACATGCGTATCGAGATACCGAACCCGGTGGCCGACTTGTATACCTTTTATGGACGCCTCGAGATCCCGGGGCTAGACAGCCTGGTGCTGTCCACCGATAACCTCATGCTTAGAGGATCCAGAGTCAAAAATACGGAGTGGGCCATAGGTTGTGCTATTTACACAGGGGAAGAAAGTAAACTGGCTCTCAATTCGAAATACACAGGGAACAAGTTCTCATCGTGCGAACTGGCCGTCAACGGGTTCCTGCTCTTCTTTATTTTGGTATTACTCATCGAGATGGTCGTGTCTTTGATCGCCAAGCTTGTCATAGAGCACAGGAACCCCGAGAAGGACCTGTACCTCGGTCCGGGCACCACGTTCCCGCCGCACGTCTCCGACCTACTTCAGGATCTTTTCTCGTTTCTGCTGCTATATTATTATATCATACCGATGTCTCTGTACGTGACCATCGAGCTTTATAAATTTATAGGAGCACTATTTATCGGATGGGACGAAGACTTTCGCTGCGAGGACACAGGAAGACCGGCGATGGCCAACACATCGGACCTCAACGAAGAGCTAGGTCAGGTCGAGGTTTTGTTCTCAGACAAAACTGGCACCTTAACGAAAAACCTCATGGTGTTCAAGACATGTTCAATCAAAGGCAACGTTTACGAAGAGAGGAGTAACCAGCTGTACAACGTGGAAAGGTTTGAAGAGCCCGTCGATGTGCTTCAGACAGACATCAAATTTTTCTTCAAAATCCTCGCCCTGTGTCATTCGGTTCAAGTTTCCAGTGAAGACATGAAGAAGCTTAGCGCACGACTGTCTGGCGGAGGAGATACTCAACTGATGAACTTCTTTCGAAGGAAAAAACAAAATAAAAATCCCAACAACGGCGTACTGGACAATGTCAGTTGGGACTCGGCCTTGAAAGAAGCTGGCAGTAAAATGGACTACCAAGGCAGCAGCCCCGATGAGAAAGCATTGGTGGAGGCCGCCGATAGAGTGGGAGTCACGTTTTTGGGCGAAGAGGGCAATAATCTCGTATTGAAAGTGGGTGAGAACACGGAAATGTATGAAAGATTGCAACTCATCGAATTCACTTCGGAGAGGAAGAGGATGTCAGTTATTCTTCGTGATAAAGACGGAAAGATATGGTTATTTTGCAAGGGAGCCGAAAGCTCTGTGTTTCCACTGTGCAAAGACTCGGCTCTGGTGGAGGAGGTGAACAGGGACATCAATGCGTTCGCAGTGAAGGGACTGCGGACATTAGCTATTGCTTACAGAGAGATTTCTGACGAGGAATACACTAGAGTCTCCGAAAGCATAAAGCATGTAGACGGAAAGAGCGCGGAGGCTCTGCAGCAACTCACCCAGCAGTACCGGGTTTTAGAACAACAACTGATATTAGTCGGCGCCACTGGAGTCGAGGACTGTCTGCAGGACGACGTGGCCGACACGCTCGCCTCCTTACGAAGAGCTGGCATCAAAACCTGGGTGCTCACTGGGGATAAGGTGGAAACAGCGCTCAACGTGGCCCAGTCGTGCGCGCACATATCGGAGACGGACCGGCGCATGTTCCTGGTCGGGGTGGAGTCGGCCGGCGCGCTGCTCGCCCACCTGGACGAGTGCCGCCGGATCGTCGCCGAGCCCGGCTACCGGGACCTCACGCTGGTGCTGGACGGGACCAGCATGACGCAGCTGCTGGACGCCGGCGCCGCGCAGGCCTTCGTCGATATATCGCTGCAGTGCAACGCCGTGCTGTGCTGCCGACTGTCGCCCATACAGAAGGCTAAGATAGTGAGACTTATAAAGAGCAGTCCGCAGCATCCGATCACGGCCGCTATAGGCGACGGCGCTAATGACATCTCTATGATACAAGAGGCGCACGTCGGCTTCGGTATATTCGGAAGAGAGGGTCATCAGGCCGCGAGATCCGCAGATTTCGCCATAACAAAATTTTCAATGATAAAAAAGTTTTTACTCGTGTTGGGACATTGGTACTATCAGCGGCTGGCGACGCTCATACACTACTTCTTCTACAAAAATCTAGTCTTAGGTAATATAATGTTCGCGTTTCAAATCCAAACGGCCTTTTCGACACAATCGGTATACGACTCGATGTACCTGACGCTGTTCAACCTGCTGTTCACGTCGCTGCCCTGCCTCATCCTGTCGGTGACGGAGCAGCGCTGGCCGGCCGACATGCTGCTCAAAAACCCGACACTGTACAGAAGCATCACGAAGAACAAGCTGATGACGTGGTCTAACTTCGGGGTGTGGTGCCTCTCGGCCGTGTACCACTCTGTTGTCATTTACATGTTCACGTATCTGATATTCGTCGTGCCCGTCATTAATCTCGACGGGAACACTGTCGACTTGTGGTCGTACGGCGCTGCGCTATTCCACCTCACGATGGTCACCGTGAGTCTCAAGCTGTGGCTGCAGGCGCGCTACCACACGCTCGTGTTCGTCGCCTCCACCCTGCTCTCCTTATTCAGCTACCTCGTCTTCAACACGATCTACTCGCTCGTCTATCTACAAATCGACGGTGACGTGCTAGGGACGTACATTCGTTTACTAACATCTCCTGCGTTCGCATTTCTAAACTTGGTGGTTGTGATCGCGACGCTAGTGCCGGACTACTGCGTGCGGCTCGCGTCGGACCGCGGGGGCCGCCCCCGCCACGCGCCCCCCACCCGCGCGCCTCCCGCCGCCCTCTCCACCAGGCTCTAG
Protein
MVSSDSPVSPFTSIAPLSFMVFVTAVKQGYEDWLRHKTDNKVNNQIVEIVYKGVIKEVKNSTIAPGTLVRVKRGREVPADLVLLCSAGEKGKCYITTANLDGETNLKTLRVPPPLVGYTPDILPQNMRIEIPNPVADLYTFYGRLEIPGLDSLVLSTDNLMLRGSRVKNTEWAIGCAIYTGEESKLALNSKYTGNKFSSCELAVNGFLLFFILVLLIEMVVSLIAKLVIEHRNPEKDLYLGPGTTFPPHVSDLLQDLFSFLLLYYYIIPMSLYVTIELYKFIGALFIGWDEDFRCEDTGRPAMANTSDLNEELGQVEVLFSDKTGTLTKNLMVFKTCSIKGNVYEERSNQLYNVERFEEPVDVLQTDIKFFFKILALCHSVQVSSEDMKKLSARLSGGGDTQLMNFFRRKKQNKNPNNGVLDNVSWDSALKEAGSKMDYQGSSPDEKALVEAADRVGVTFLGEEGNNLVLKVGENTEMYERLQLIEFTSERKRMSVILRDKDGKIWLFCKGAESSVFPLCKDSALVEEVNRDINAFAVKGLRTLAIAYREISDEEYTRVSESIKHVDGKSAEALQQLTQQYRVLEQQLILVGATGVEDCLQDDVADTLASLRRAGIKTWVLTGDKVETALNVAQSCAHISETDRRMFLVGVESAGALLAHLDECRRIVAEPGYRDLTLVLDGTSMTQLLDAGAAQAFVDISLQCNAVLCCRLSPIQKAKIVRLIKSSPQHPITAAIGDGANDISMIQEAHVGFGIFGREGHQAARSADFAITKFSMIKKFLLVLGHWYYQRLATLIHYFFYKNLVLGNIMFAFQIQTAFSTQSVYDSMYLTLFNLLFTSLPCLILSVTEQRWPADMLLKNPTLYRSITKNKLMTWSNFGVWCLSAVYHSVVIYMFTYLIFVVPVINLDGNTVDLWSYGAALFHLTMVTVSLKLWLQARYHTLVFVASTLLSLFSYLVFNTIYSLVYLQIDGDVLGTYIRLLTSPAFAFLNLVVVIATLVPDYCVRLASDRGGRPRHAPPTRAPPAALSTRL
Summary
Catalytic Activity
ATP + H(2)O + phospholipid(Side 1) = ADP + phosphate + phospholipid(Side 2).
Similarity
Belongs to the cation transport ATPase (P-type) (TC 3.A.3) family. Type IV subfamily.
Feature
chain Phospholipid-transporting ATPase
Uniprot
H9J8N8
A0A2A4JGX5
A0A2A4JI17
A0A0N1ICK3
A0A212EYA4
A0A3S2TLV9
+ More
A0A2H1V7H0 A0A194PTX9 A0A0L7LJX0 A0A3L8DPM6 A0A1I8MMY2 A0A158NBC3 A0A154P2L5 A0A195BRN2 A0A0P5CUR1 E9IZZ4 A0A0P6I834 A0A195EVS2 A0A0P5VVB4 A0A164X0T1 A0A310SM57 A0A151WZT7 A0A0P5TKU8 A0A0P5F034 E9G9U0 A0A087ZVH1 A0A1D1V773 A0A0P6H619 A0A0N8BD34 A0A0P6FUS5 A0A0P5LFJ1 A0A0P5JQ22 A0A0P6DMG9 A0A0P6BQ31 A0A164X0Q0 A0A0P6DR54 A0A0P5N9Q8 A0A0N8E883 A0A0P5MRH2 A0A0P6I126 A0A0P5N3N6 K7FGF8 A0A0P7THG5 A0A2K6ENH8 A0A1W4Y569 A0A232FIS1 A0A3Q2GJS7 A0A3Q2GJS3 A0A1W4YE45 K7J7V1 A0A1W4YEF9 A0A1W4YEG5 A0A3Q3M7I3 A0A0D9RL82 A0A091JND1 A0A091SLJ8 A0A2K5Q2X0 A0A2K5Q2T3 A0A1A7WIW2 A0A2K5Q2Z1 S7P0T6 A0A287DDK9 A0A2K5Q2Y0 A0A3Q3LZX4 A0A1D5NT64 I3MRL2 A0A250Y702 U6DGU8 A0A093I441 A0A287CYN6 F6Y2P4 A0A2J8WGY7 A0A3Q7PJL7 A0A0P6G427 A0A091VTE9 A0A2K6RIL2 A0A0P6HJJ5 A0A3Q1GIY6 F1NFH6 L5JP87 A0A2K6RIN7 A0A1U7UFF9 A0A2U3ZN90 A0A0P5Z2B5 A0A3B5B8C0 A0A151M6B0 A0A091WMN0 A0A3Q1KD09 A0A1S3GJW1 A0A2Y9JKJ2 M3XTD4 A0A2K6E928 A0A2K6A5V4 A0A2R9C3I1 A0A2I3SUK6 A0A2U3WE60 A0A2K5HXG7 A0A091HL09 A0A091M987
A0A2H1V7H0 A0A194PTX9 A0A0L7LJX0 A0A3L8DPM6 A0A1I8MMY2 A0A158NBC3 A0A154P2L5 A0A195BRN2 A0A0P5CUR1 E9IZZ4 A0A0P6I834 A0A195EVS2 A0A0P5VVB4 A0A164X0T1 A0A310SM57 A0A151WZT7 A0A0P5TKU8 A0A0P5F034 E9G9U0 A0A087ZVH1 A0A1D1V773 A0A0P6H619 A0A0N8BD34 A0A0P6FUS5 A0A0P5LFJ1 A0A0P5JQ22 A0A0P6DMG9 A0A0P6BQ31 A0A164X0Q0 A0A0P6DR54 A0A0P5N9Q8 A0A0N8E883 A0A0P5MRH2 A0A0P6I126 A0A0P5N3N6 K7FGF8 A0A0P7THG5 A0A2K6ENH8 A0A1W4Y569 A0A232FIS1 A0A3Q2GJS7 A0A3Q2GJS3 A0A1W4YE45 K7J7V1 A0A1W4YEF9 A0A1W4YEG5 A0A3Q3M7I3 A0A0D9RL82 A0A091JND1 A0A091SLJ8 A0A2K5Q2X0 A0A2K5Q2T3 A0A1A7WIW2 A0A2K5Q2Z1 S7P0T6 A0A287DDK9 A0A2K5Q2Y0 A0A3Q3LZX4 A0A1D5NT64 I3MRL2 A0A250Y702 U6DGU8 A0A093I441 A0A287CYN6 F6Y2P4 A0A2J8WGY7 A0A3Q7PJL7 A0A0P6G427 A0A091VTE9 A0A2K6RIL2 A0A0P6HJJ5 A0A3Q1GIY6 F1NFH6 L5JP87 A0A2K6RIN7 A0A1U7UFF9 A0A2U3ZN90 A0A0P5Z2B5 A0A3B5B8C0 A0A151M6B0 A0A091WMN0 A0A3Q1KD09 A0A1S3GJW1 A0A2Y9JKJ2 M3XTD4 A0A2K6E928 A0A2K6A5V4 A0A2R9C3I1 A0A2I3SUK6 A0A2U3WE60 A0A2K5HXG7 A0A091HL09 A0A091M987
EC Number
7.6.2.1
Pubmed
EMBL
BABH01030370
BABH01030371
NWSH01001496
PCG71069.1
PCG71070.1
KQ460954
+ More
KPJ10254.1 AGBW02011576 OWR46472.1 RSAL01000060 RVE49642.1 ODYU01000832 SOQ36194.1 KQ459592 KPI96871.1 JTDY01000838 KOB75715.1 QOIP01000006 RLU22183.1 ADTU01010853 ADTU01010854 ADTU01010855 KQ434804 KZC06097.1 KQ976417 KYM89670.1 GDIP01181463 JAJ41939.1 GL767291 EFZ13815.1 GDIQ01009216 JAN85521.1 KQ981953 KYN32338.1 GDIP01094734 JAM08981.1 LRGB01001036 KZS13759.1 KQ762207 OAD56072.1 KQ982630 KYQ53419.1 GDIP01126472 JAL77242.1 GDIP01153322 JAJ70080.1 GL732536 EFX83825.1 BDGG01000002 GAU94683.1 GDIQ01130269 GDIQ01033905 JAN60832.1 GDIQ01189587 JAK62138.1 GDIQ01044106 JAN50631.1 GDIQ01170353 JAK81372.1 GDIQ01195584 GDIQ01139058 GDIQ01130268 JAK56141.1 GDIQ01074785 JAN19952.1 GDIP01011466 JAM92249.1 KZS13758.1 GDIQ01074784 JAN19953.1 GDIQ01152304 JAK99421.1 GDIQ01052204 JAN42533.1 GDIQ01152305 JAK99420.1 GDIQ01018874 JAN75863.1 GDIQ01155157 JAK96568.1 AGCU01198561 AGCU01198562 AGCU01198563 AGCU01198564 AGCU01198565 AGCU01198566 JARO02010907 KPP60303.1 NNAY01000185 OXU30197.1 AAZX01001793 AQIB01119177 AQIB01119178 KK501163 KFP13201.1 KK477793 KFQ59527.1 HADW01004270 SBP05670.1 KE161377 EPQ03498.1 AGTP01061711 AGTP01061712 AGTP01061713 AADN05000526 GFFV01000533 JAV39412.1 HAAF01009997 CCP81821.1 KL206809 KFV85952.1 AAEX03017263 AAEX03017264 NDHI03003390 PNJ69033.1 GDIQ01139059 GDIQ01038799 JAN55938.1 KL411359 KFR06025.1 GDIQ01032109 JAN62628.1 KB031155 ELK00742.1 GDIP01051036 JAM52679.1 AKHW03006437 KYO20038.1 KK735892 KFR16375.1 AEYP01017042 AEYP01017043 AEYP01017044 AEYP01017045 AEYP01017046 AEYP01017047 AJFE02113381 AJFE02113382 AACZ04067283 KL515867 KFO88003.1 KK526564 KFP68343.1
KPJ10254.1 AGBW02011576 OWR46472.1 RSAL01000060 RVE49642.1 ODYU01000832 SOQ36194.1 KQ459592 KPI96871.1 JTDY01000838 KOB75715.1 QOIP01000006 RLU22183.1 ADTU01010853 ADTU01010854 ADTU01010855 KQ434804 KZC06097.1 KQ976417 KYM89670.1 GDIP01181463 JAJ41939.1 GL767291 EFZ13815.1 GDIQ01009216 JAN85521.1 KQ981953 KYN32338.1 GDIP01094734 JAM08981.1 LRGB01001036 KZS13759.1 KQ762207 OAD56072.1 KQ982630 KYQ53419.1 GDIP01126472 JAL77242.1 GDIP01153322 JAJ70080.1 GL732536 EFX83825.1 BDGG01000002 GAU94683.1 GDIQ01130269 GDIQ01033905 JAN60832.1 GDIQ01189587 JAK62138.1 GDIQ01044106 JAN50631.1 GDIQ01170353 JAK81372.1 GDIQ01195584 GDIQ01139058 GDIQ01130268 JAK56141.1 GDIQ01074785 JAN19952.1 GDIP01011466 JAM92249.1 KZS13758.1 GDIQ01074784 JAN19953.1 GDIQ01152304 JAK99421.1 GDIQ01052204 JAN42533.1 GDIQ01152305 JAK99420.1 GDIQ01018874 JAN75863.1 GDIQ01155157 JAK96568.1 AGCU01198561 AGCU01198562 AGCU01198563 AGCU01198564 AGCU01198565 AGCU01198566 JARO02010907 KPP60303.1 NNAY01000185 OXU30197.1 AAZX01001793 AQIB01119177 AQIB01119178 KK501163 KFP13201.1 KK477793 KFQ59527.1 HADW01004270 SBP05670.1 KE161377 EPQ03498.1 AGTP01061711 AGTP01061712 AGTP01061713 AADN05000526 GFFV01000533 JAV39412.1 HAAF01009997 CCP81821.1 KL206809 KFV85952.1 AAEX03017263 AAEX03017264 NDHI03003390 PNJ69033.1 GDIQ01139059 GDIQ01038799 JAN55938.1 KL411359 KFR06025.1 GDIQ01032109 JAN62628.1 KB031155 ELK00742.1 GDIP01051036 JAM52679.1 AKHW03006437 KYO20038.1 KK735892 KFR16375.1 AEYP01017042 AEYP01017043 AEYP01017044 AEYP01017045 AEYP01017046 AEYP01017047 AJFE02113381 AJFE02113382 AACZ04067283 KL515867 KFO88003.1 KK526564 KFP68343.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000283053
UP000053268
+ More
UP000037510 UP000279307 UP000095301 UP000005205 UP000076502 UP000078540 UP000078541 UP000076858 UP000075809 UP000000305 UP000005203 UP000186922 UP000007267 UP000034805 UP000233160 UP000192224 UP000215335 UP000265020 UP000002358 UP000261640 UP000029965 UP000053119 UP000233040 UP000005215 UP000000539 UP000053584 UP000002254 UP000286641 UP000053283 UP000233200 UP000257200 UP000010552 UP000189704 UP000245340 UP000261400 UP000050525 UP000053605 UP000265040 UP000081671 UP000248482 UP000000715 UP000233120 UP000233140 UP000240080 UP000002277 UP000233080
UP000037510 UP000279307 UP000095301 UP000005205 UP000076502 UP000078540 UP000078541 UP000076858 UP000075809 UP000000305 UP000005203 UP000186922 UP000007267 UP000034805 UP000233160 UP000192224 UP000215335 UP000265020 UP000002358 UP000261640 UP000029965 UP000053119 UP000233040 UP000005215 UP000000539 UP000053584 UP000002254 UP000286641 UP000053283 UP000233200 UP000257200 UP000010552 UP000189704 UP000245340 UP000261400 UP000050525 UP000053605 UP000265040 UP000081671 UP000248482 UP000000715 UP000233120 UP000233140 UP000240080 UP000002277 UP000233080
PRIDE
Interpro
IPR036412
HAD-like_sf
+ More
IPR023214 HAD_sf
IPR008250 ATPase_P-typ_transduc_dom_A_sf
IPR023298 ATPase_P-typ_TM_dom_sf
IPR001757 P_typ_ATPase
IPR023299 ATPase_P-typ_cyto_dom_N
IPR006539 P-type_ATPase_IV
IPR032630 P_typ_ATPase_c
IPR018303 ATPase_P-typ_P_site
IPR032631 P-type_ATPase_N
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR030362 ATP11B
IPR023214 HAD_sf
IPR008250 ATPase_P-typ_transduc_dom_A_sf
IPR023298 ATPase_P-typ_TM_dom_sf
IPR001757 P_typ_ATPase
IPR023299 ATPase_P-typ_cyto_dom_N
IPR006539 P-type_ATPase_IV
IPR032630 P_typ_ATPase_c
IPR018303 ATPase_P-typ_P_site
IPR032631 P-type_ATPase_N
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR030362 ATP11B
SUPFAM
Gene 3D
ProteinModelPortal
H9J8N8
A0A2A4JGX5
A0A2A4JI17
A0A0N1ICK3
A0A212EYA4
A0A3S2TLV9
+ More
A0A2H1V7H0 A0A194PTX9 A0A0L7LJX0 A0A3L8DPM6 A0A1I8MMY2 A0A158NBC3 A0A154P2L5 A0A195BRN2 A0A0P5CUR1 E9IZZ4 A0A0P6I834 A0A195EVS2 A0A0P5VVB4 A0A164X0T1 A0A310SM57 A0A151WZT7 A0A0P5TKU8 A0A0P5F034 E9G9U0 A0A087ZVH1 A0A1D1V773 A0A0P6H619 A0A0N8BD34 A0A0P6FUS5 A0A0P5LFJ1 A0A0P5JQ22 A0A0P6DMG9 A0A0P6BQ31 A0A164X0Q0 A0A0P6DR54 A0A0P5N9Q8 A0A0N8E883 A0A0P5MRH2 A0A0P6I126 A0A0P5N3N6 K7FGF8 A0A0P7THG5 A0A2K6ENH8 A0A1W4Y569 A0A232FIS1 A0A3Q2GJS7 A0A3Q2GJS3 A0A1W4YE45 K7J7V1 A0A1W4YEF9 A0A1W4YEG5 A0A3Q3M7I3 A0A0D9RL82 A0A091JND1 A0A091SLJ8 A0A2K5Q2X0 A0A2K5Q2T3 A0A1A7WIW2 A0A2K5Q2Z1 S7P0T6 A0A287DDK9 A0A2K5Q2Y0 A0A3Q3LZX4 A0A1D5NT64 I3MRL2 A0A250Y702 U6DGU8 A0A093I441 A0A287CYN6 F6Y2P4 A0A2J8WGY7 A0A3Q7PJL7 A0A0P6G427 A0A091VTE9 A0A2K6RIL2 A0A0P6HJJ5 A0A3Q1GIY6 F1NFH6 L5JP87 A0A2K6RIN7 A0A1U7UFF9 A0A2U3ZN90 A0A0P5Z2B5 A0A3B5B8C0 A0A151M6B0 A0A091WMN0 A0A3Q1KD09 A0A1S3GJW1 A0A2Y9JKJ2 M3XTD4 A0A2K6E928 A0A2K6A5V4 A0A2R9C3I1 A0A2I3SUK6 A0A2U3WE60 A0A2K5HXG7 A0A091HL09 A0A091M987
A0A2H1V7H0 A0A194PTX9 A0A0L7LJX0 A0A3L8DPM6 A0A1I8MMY2 A0A158NBC3 A0A154P2L5 A0A195BRN2 A0A0P5CUR1 E9IZZ4 A0A0P6I834 A0A195EVS2 A0A0P5VVB4 A0A164X0T1 A0A310SM57 A0A151WZT7 A0A0P5TKU8 A0A0P5F034 E9G9U0 A0A087ZVH1 A0A1D1V773 A0A0P6H619 A0A0N8BD34 A0A0P6FUS5 A0A0P5LFJ1 A0A0P5JQ22 A0A0P6DMG9 A0A0P6BQ31 A0A164X0Q0 A0A0P6DR54 A0A0P5N9Q8 A0A0N8E883 A0A0P5MRH2 A0A0P6I126 A0A0P5N3N6 K7FGF8 A0A0P7THG5 A0A2K6ENH8 A0A1W4Y569 A0A232FIS1 A0A3Q2GJS7 A0A3Q2GJS3 A0A1W4YE45 K7J7V1 A0A1W4YEF9 A0A1W4YEG5 A0A3Q3M7I3 A0A0D9RL82 A0A091JND1 A0A091SLJ8 A0A2K5Q2X0 A0A2K5Q2T3 A0A1A7WIW2 A0A2K5Q2Z1 S7P0T6 A0A287DDK9 A0A2K5Q2Y0 A0A3Q3LZX4 A0A1D5NT64 I3MRL2 A0A250Y702 U6DGU8 A0A093I441 A0A287CYN6 F6Y2P4 A0A2J8WGY7 A0A3Q7PJL7 A0A0P6G427 A0A091VTE9 A0A2K6RIL2 A0A0P6HJJ5 A0A3Q1GIY6 F1NFH6 L5JP87 A0A2K6RIN7 A0A1U7UFF9 A0A2U3ZN90 A0A0P5Z2B5 A0A3B5B8C0 A0A151M6B0 A0A091WMN0 A0A3Q1KD09 A0A1S3GJW1 A0A2Y9JKJ2 M3XTD4 A0A2K6E928 A0A2K6A5V4 A0A2R9C3I1 A0A2I3SUK6 A0A2U3WE60 A0A2K5HXG7 A0A091HL09 A0A091M987
PDB
6A69
E-value=3.30497e-14,
Score=195
Ontologies
GO
PANTHER
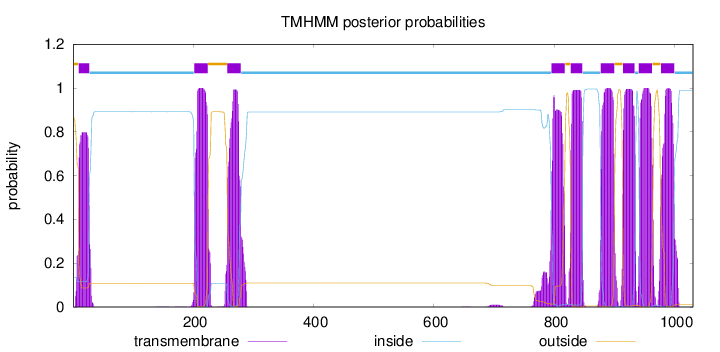
Topology
Subcellular location
Membrane
Length:
1031
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
192.95372
Exp number, first 60 AAs:
15.49642
Total prob of N-in:
0.13639
POSSIBLE N-term signal
sequence
outside
1 - 9
TMhelix
10 - 27
inside
28 - 201
TMhelix
202 - 224
outside
225 - 256
TMhelix
257 - 279
inside
280 - 795
TMhelix
796 - 818
outside
819 - 827
TMhelix
828 - 847
inside
848 - 877
TMhelix
878 - 900
outside
901 - 914
TMhelix
915 - 934
inside
935 - 940
TMhelix
941 - 963
outside
964 - 977
TMhelix
978 - 1000
inside
1001 - 1031
Population Genetic Test Statistics
Pi
18.400294
Theta
17.140457
Tajima's D
-1.14336
CLR
0.735761
CSRT
0.113394330283486
Interpretation
Uncertain
